Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Since its first report in Wuhan, China, in December 2019, COVID-19 has become a pandemic, affecting millions of people worldwide. Although the virus primarily affects the respiratory tract, gastrointestinal symptoms are also common.
- COVID-19
- SARS-CoV-2
- gastrointestinal tract
1. Introduction
Since the first report in December 2019 in Wuhan, China, coronavirus disease 2019 (COVID-19) spread all over the world, causing more than 758 million cases and 6.85 million deaths [1].
The clinical course of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infection ranges from asymptomatic to a rapidly progressing and life-threatening disease and is associated with a variety of symptoms [2].
Like other coronaviruses, SARS-CoV-2 infects the gastrointestinal tract, inducing nausea, vomiting, abdominal pain, and diarrhea [3][4][5].
Since the COVID-19 occurrence in late 2019, intense research efforts on an unprecedented scale have focused on the study of SARS-CoV-2 entry mechanisms and clinical presentations.
Prior to COVID-19, there were two short-lived pandemics—SARS-CoV-1 in 2002 and MERS in 2012. The first cases of SARS-CoV-1 were detected in China and quickly spread with a high lethality rate of 11%, resulting in 8422 reported cases and 916 deaths. Similarly, MERS emerged in Saudi Arabia with a high mortality rate of approximately 37% and was also traced back to bats. Both viruses caused similar symptoms such as fever, cough, dyspnea, and atypical pneumonia, as well as affecting the gastrointestinal tract with diarrhea being a common symptom. However, MERS had a higher prevalence of GI symptoms, mortality rate, and need for extreme treatment measures such as mechanical ventilation compared to SARS-CoV-1. Despite the potential for fecal–oral transmission, extrapulmonary symptoms were not given much attention due to the short-lived and localized nature of these pandemics [6][7].
2. Prevalence of Gastrointestinal Symptoms
Several studies reported gastrointestinal symptoms in patients affected by COVID-19. Their prevalence in adults is high, with diarrhea, nausea, and abdominal pain being the most frequent ones (16.5%, 9.7%, 4.5%, respectively) [8]. Anorexia or loss of appetite (1.6%), vomiting (1.5%), and loss of taste (1.3%) are less common. Increased liver enzymes are not rare (5.6%) (Table 1) [8]. Prevalences, however, are hardly comparable as different series do not report all main gastrointestinal symptoms. This likely results from the different weight attributed by authors to mild and/or infrequent symptoms, which were not invariably reported.
Table 1. Gastrointestinal symptoms in adult and pediatric patients.
| Adult Patients | Pediatric Patients | |
|---|---|---|
| Diarrhea | 16.5% | 19.0% |
| Nausea/vomiting | 11.2% | 19.7% |
| abdominal pain | 4.5% | 20.3% |
| Anorexia | 1.6% | 10% |
In the pediatric population, the incidence of gastrointestinal symptoms is higher compared to adults [9]. A recent meta-analysis reported a higher prevalence of nausea/vomiting (19.7%) and abdominal pain (20.3%) but not diarrhea (19.08%) [9]. However, wide variations have been described in different series [10].
The relationship between gastrointestinal symptoms, mortality, and more severe systemic disease presentation is debatable. Available evidence suggests that the overall presence of gastrointestinal symptoms is not associated with increased mortality rates (OR = 0.88; 95% CI 0.71–1.09; p = 0.23). The same applies to individual symptoms, such as diarrhea (p = 0.96), nausea/vomiting (p = 0.46), and abdominal pain (p = 0.3) [11].
A recent meta-analysis correlating the incidence of gastrointestinal symptoms with severe presentation in adults showed that abdominal pain (OR = 2.70, 95% CI 1.17–6.27, p = 0.02), but not diarrhea, nausea, or vomiting, is associated with aggressive disease [12].
In the pediatric population, the presence of diarrhea significantly correlates with more severe clinical course (OR 3.9, 95% CI: 1.80–8.73; p < 0.01). This is not the case for nausea/vomiting and abdominal pain [9].
The persistence of gastrointestinal symptoms for more than two weeks after discharge is also high, as 3.23% of patients reported nausea, 3.19% vomiting, and 4.12% prolonged diarrhea, while the persistence of abdominal pain was 1.68% [13].
Gastrointestinal bleeding (GIB) was reported in 2% of COVID-19 patients, respectively, 1% for upper GI and 1% for lower GI [14]. The most common causes were ulcers (25.3%), erosive/ulcerative diffuse damage (16.1%), and petechial/hemorrhagic gastropathy (9.2%) in upper gastrointestinal bleeding. Ischemic colitis was reported in one-third of patients with lower gastrointestinal bleeding [15]. Again, some differences have been reported in different cohorts [4]. The presentation with GIB however represents a negative prognostic factor, as reported by a recent systematic review comparing the outcomes of 808 COVID-19 patients with GIB and 18,179 non-GIB COVID-19 patients. The overall incidence of GIB was 0.06%, with GIB patients showing a higher death rate than non-GIB patients (25.4% vs. 16.4%, p < 0.001) [4].
3. Gastrointestinal Infection
Respiratory droplets are the main route of transmission of SARS-CoV-2 [16], but SARS-CoV-2 RNA was also detected in fecal samples [17][18][19][20].
Direct evidence of fecal–oral transmission is still lacking, but emerging evidence supports the hypothesis [21][22]. SARS-CoV-2 RNA has indeed been detected in anal swabs and stool samples in over 50% of infected patients [23][24][25]. RNA levels in the stools ranged from 102 to 105 copies/mL, but in several reports fecal shedding exceeded 107 copies/mL [23][26][27][28]. This is in line with nasopharyngeal fluids concentration (105–1011 copies/mL) [27][28]. SARS-CoV-2 concentration in stool samples peaks 2 to 3 weeks after symptom onset [25][29], and, as reported in a small German cohort, the RNA load in fecal specimens reflects what is found in sputum in 86% of cases (6 of 7 patients) [23]. Live SARS-CoV-2 was also observed in the feces of COVID-19 patients, confirming potential fecal–oral transmission [30]. However, despite being easily detected by electron microscopy [31], SARS-CoV-2 isolation from stools is difficult [32].
Recent evidence suggests that viral variants show different gastrointestinal infectivity. Reduced viral replication of Omicron BA.1 and BA.2 variants compared with the B.1.617.2/Delta variant was reported in studies carried out in organoids [33].
The persistence of the virus in stools is significantly longer (median 22 days, interquartile range 17–31 days) than in respiratory (18 days, 13–29 days; p = 0.02) and serum samples (16 days, 11–21 days; p < 0.001) [25]. Polymerase chain reaction detected viral RNA in fecal samples of patients with no detectable virus in respiratory tract specimens [19][25][34].
Several studies have shown the presence of SARS-CoV-2 in epithelial cells of both the small and large intestines, as demonstrated by intestinal biopsies [17][35][36]. Additionally, the transcription of subgenomic SARS-CoV-2 mRNA (sgmRNA) indicates active viral replication in the intestine, as sgmRNA is only transcribed in infected cells and not packaged into virions [23]. Intestinal organoids primarily secrete SARS-CoV-2 apically [37], which may explain viral excretion in feces.
Although evidence suggests that the gut is an active site of SARS-CoV-2 replication, it is still unclear whether the virus present in feces is directly infectious. While viral RNA may be shed in fecal specimens, the presence of viral ribonucleic acid does not necessarily imply the presence of live transmissible virus [38]. Therefore, while direct and indirect data support the hypothesis that SARS-CoV-2 actively infects human intestinal epithelial cells, further research is needed to determine whether the virus in feces is directly infectious.
4. SARS-CoV-2 Structure and Interaction with the Host
SARS-CoV-2 is a single-stranded β-coronavirus, with 29.9 kb RNA genome and an envelope with spikes on the surface (Figure 1) [39]. It shares up to 80% of the gene sequence with other pathogenic members of the coronavirus family, such as SARS-CoV-1 and Middle East Respiratory Syndrome coronavirus (MERS-CoV) [40].
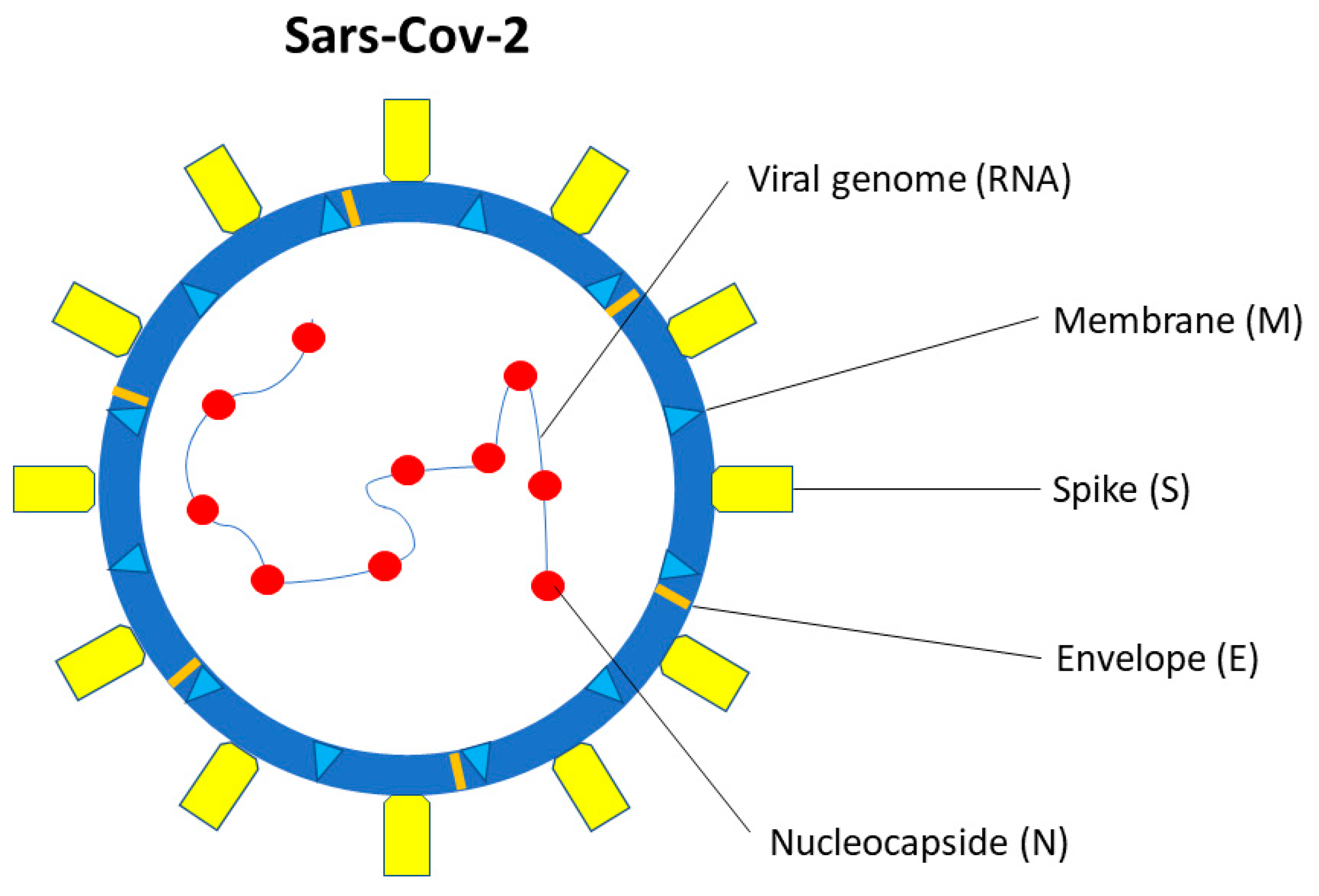
Figure 1. SARS-CoV-2 structure. SARS-CoV-2 is a single-stranded β-coronavirus. Two-thirds of viral RNA, in the first open reading frame (ORF 1a/b), encodes for 16 non-structural proteins (NSP). The remaining viral genome expresses accessory proteins, interfering with the host innate immune response, and four structural proteins (S, E, N, M).
Two-thirds of viral RNA, in the first open reading frame (ORF 1a/b), translates two polyproteins, encoding for 16 non-structural proteins (NSP). The remaining viral genome expresses accessory proteins, interfering with the host innate immune response, and structural proteins. The four essential structural proteins include the spike (S) glycoprotein, small envelope (E) protein, nucleocapsid (N) protein, and matrix (M) protein [41]. Proteins M, E, and S form the viral envelope [42]. The N protein is structurally bound to the viral RNA and is involved in viral replication [43], while the M protein plays a role in determining the shape of the viral envelope [43].
Open reading frames encoding nine accessory proteins (3a, 3b, 6, 7a, 7b, 8, 9b, 9c, and 10) and two polyproteins (pp1a and pp1ab) are also present in the genome [44].
Polyproteins pp1a and pp1ab, but not accessory proteins, are involved in viral replication [44].
SARS-CoV-2 interacts with target cells through envelope spike glycoprotein which binds the angiotensin-converting enzyme 2 (ACE2) receptor of the host (Figure 2) [41]. The binding affinity for the human ACE2 receptor is 10–20 times stronger for SARS-CoV-2 than for its predecessor SARS-CoV-1 [45]. After binding ACE2 receptors, the transmembrane serine protease 2 (TMPRSS2) mediates the cleavage of the spike glycoprotein, regulating the internalization of the virus into target cells [46]. The two subunits, S1 and S2, favor the binding of the virus to the cell and the fusion between the two cellular membranes [46].
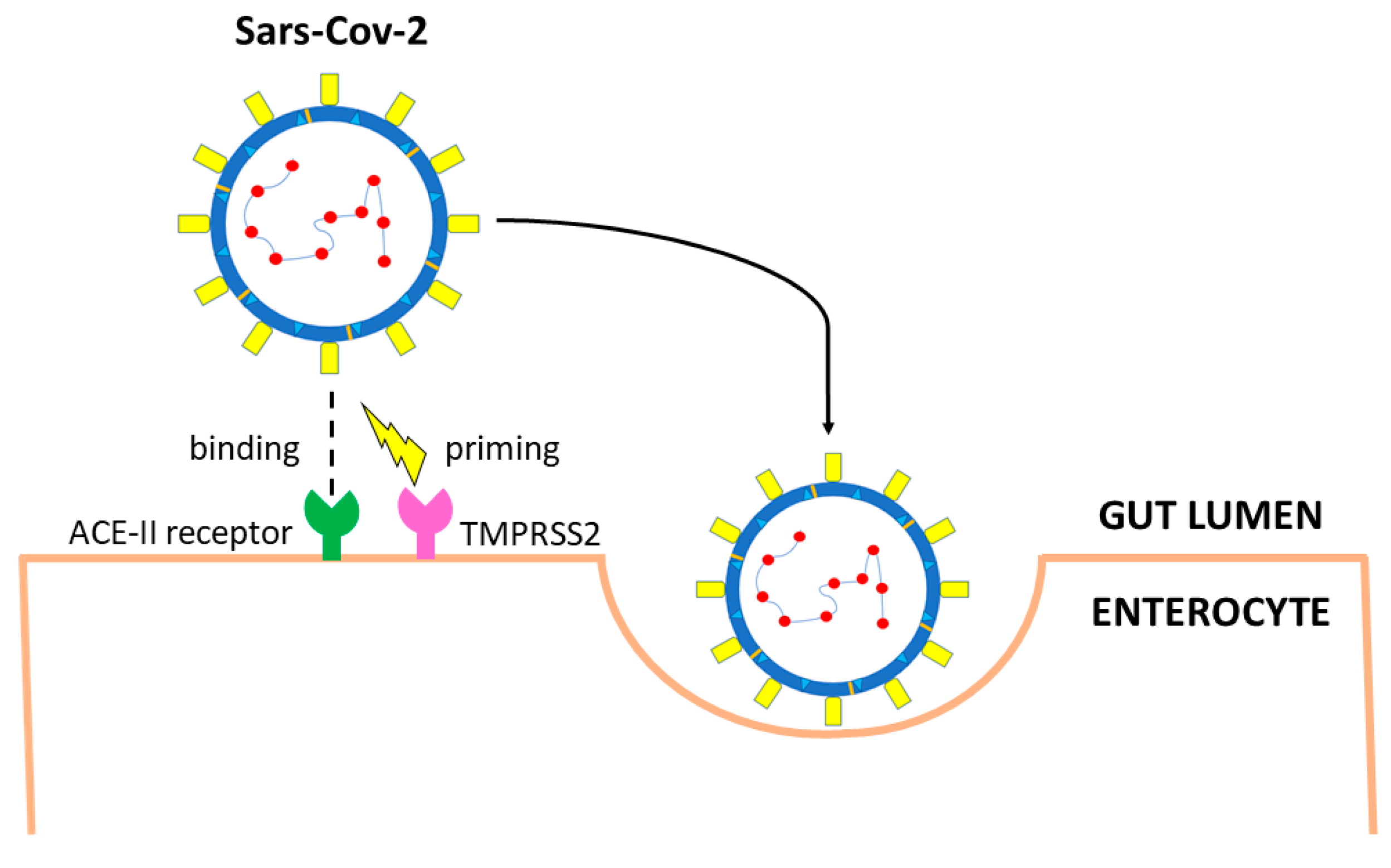
Figure 2. Main entry route of SARS-CoV-2 in gastrointestinal tract. The spike protein of SARS-CoV-2 binds angiotensin-converting enzyme 2 (ACE2) receptors. Priming by TMPRSS2, TMPRSS4 (or other proteases such as FURIN) leads to endocytosis. The virus is uncoated, genomic RNA is released, and viral proteins are synthesized using the host replication system.
Two proteolytic events are however needed to activate SARS-CoV-2. The first is the cut in the specific cleavage site between the S1 and S2 domains, which is recognized by various proteases, including TMPRSS2 [46] and furin [47][48]. The second cleavage site is within the S2 domain and allows the exposition of the fusion peptide, which enables membrane fusion. This second cleavage can be either performed by TMPRSS2 on the surface of the host cell or by lysosomal proteases, such as cathepsin L in the endolysosomes [49]. Following the entry into the cell, the virus is uncoated and replicates using the host replication system [42].
5. Route of Gastrointestinal Infection
The exact route of infection is still undefined, but the virus likely reaches the gut after being swallowed.
In vitro studies show that SARS-CoV-2, like other enveloped viruses, loses infectivity after 10 min incubation in gastric fluid [50]. Nonetheless, this route of transmission is possible as other viruses, such as influenza virus, despite being vulnerable to digestive juices, retain infectivity when protected by highly viscous mucus [51].
The glycosylation of the S protein is a further mechanism by which other coronaviruses survive the adverse milieu of the stomach and bile-salt-containing duodenal juice [52]. The mechanism may be shared by SARS-CoV-2.
Indirect evidence deriving from in vitro models of other coronaviruses suggests that fasting or fed state modulate infectivity. Indeed, MERS rapidly loses infectivity in simulated fasting state, low pH gastric fluid, but not after 2 h of exposure to fed-state condition [53].
It has also been hypothesized that chronic H. pylori infection leading to atrophic gastritis and intestinal metaplasia could facilitate SARS-CoV-2 intestinal infection by reducing stomach acidity [54][55]. H. pylori also increases the expression of ACE-2 receptors in the GI tract [56]. Clinically, a strong correlation between the occurrence of abdominal pain (19.4% vs. 2.6%, p = 0.007) or diarrhea (32.3% vs. 9.1%, p = 0.006) and H. pylori infection has been reported in COVID-19 patients [57].
Similarly, proton pump inhibitor-induced hypochloridria could favor SARS-CoV-2 intestinal infection [58], but available data are conflicting. Patients on PPI had significantly higher requirement for oxygen therapy, intensive care unit admission, and invasive ventilation than patients not taking PPI (fully adjusted OR (aOR): 2.39; 95% CI: 1.08–5.10) in a post hoc analysis from a nationwide Korean cohort [5]. Conversely, several meta-analyses did not confirm these early reports [59][60].
6. Gastrointestinal Tract–Virus Interaction
Lungs are the primary route of SARS-CoV-2 infection, but ACE2 receptors, an 805 amino acids-, type I cell-surface glycoprotein [61], are highly expressed also on the brush border of the enterocytes [62][63]. The ACE2 receptor is detected by immunofluorescent staining also in the glandular cells of the stomach and colon [17]. Viral RNA has been detected also in the esophageal mucosa, but the lack of viral nucleocapsid protein staining suggests a low viral load [17]. This is in keeping with low ACE2 expression in squamous esophageal epithelial cells. ACE2 receptors are minimally expressed by enteroendocrine cells, Paneth cells, and goblet cells [61][64][65][66].
Under physiological conditions, ACE2 receptors in the gastrointestinal tract are associated with the amino acid carrier B0AT1, which regulates the homeostasis of tryptophan, and thus stimulates the production of mechanistic target of rapamycin (mTOR)-dependent antimicrobial peptides from Paneth cells [67][68].
Besides ACE2 receptors, other molecules may play a role in SARS-CoV-2 infection. It has been demonstrated that TMPRSS2 is highly expressed in the gastrointestinal tract [50], not only in enterocytes [50] but also in intestinal goblet [69] and Paneth cells (Figure 2) [70][71]. Other serine proteases of the same family, such as TMPRSS4, are also highly expressed in mature enterocytes and potentially favor SARS-CoV-2 infection [50]. An additive effect of the two enzymes has been documented, suggesting synergic effect resulting from distinct cellular and subcellular localization of the two proteases [50].
The protease furin, widely present in the stomach, small bowel, and colon [72], also enables the S protein to separate into two pinching structures [73]. Although furin significantly increases the cleavage of the S protein, promoting SARS-CoV-2 infectivity and spread, its presence is not essential [74].
In addition to proteases, other alternative entry molecules, such as neuropilin-1 (NRP1), have been identified [75]. The expression of NRP1 in the small bowel has been reported on both human biopsies and organoids [76][77]. Following protease cleavage of the S protein into S1 and S2, a polybasic Arg-Arg-Ala-Arg carboxyl-terminal sequence on S1 binds NRP1 [75][78]. The role of NRP1 is less clear than ACE2 receptors, but the protein might mediate SARS-CoV-2 infection in ACE2-negative cells [77] and possibly explain different disease behavior in adults and children [76].
Recent studies suggest a possible interaction between the SARS-CoV-2 S protein and the cluster of differentiation (CD) 147 binding site [79], possibly through cyclophilin A-mediated regulation of ACE2 receptors [80], but conflicting results have also been published [81].
A small Italian case series [82] suggested that VEGF through CD147 may trigger gastrointestinal ischemia in SARS-CoV-2, as reported in other conditions [83]. However, validation in larger series is needed.
Protease-mediated membrane fusion represents the usual SARS-CoV-2 entry route in the host cell. An alternative option has recently been advocated, consisting of endocytosis resulting from SARS-CoV-2 binding to ACE2 receptors followed by S protein cleavage by cathepsin L [49] expressed in a variety of tissues including the gastrointestinal tract [84][85].
This entry is adapted from the peer-reviewed paper 10.3390/medicina59101709
References
- World Health Organization. Coronavirus Disease (COVID-19). Available online: https://covid19.who.int/ (accessed on 4 March 2023).
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062.
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506.
- Ashktorab, H.; Russo, T.; Oskrochi, G.; Latella, G.; Massironi, S.; Luca, M.; Chirumamilla, L.G.; Laiyemo, A.O.; Brim, H. Clinical and Endoscopic Outcomes in Coronavirus Disease-2019 Patients with Gastrointestinal Bleeding. Gastro Hep. Adv. 2022, 1, 487–499.
- Pizuorno, A.; Brim, H.; Ashktorab, H. Gastrointestinal manifestations and SARS-CoV-2 infection. Curr. Opin. Pharmacol. 2021, 61, 114–119.
- Dyall, J.; Gross, R.; Kindrachuk, J.; Johnson, R.F.; Olinger, G.G., Jr.; Hensley, L.E.; Frieman, M.B.; Jahrling, P.B. Middle East Respiratory Syndrome and Severe Acute Respiratory Syndrome: Current Therapeutic Options and Potential Targets for Novel Therapies. Drugs 2017, 77, 1935–1966.
- de Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534.
- Shehab, M.; Alrashed, F.; Shuaibi, S.; Alajmi, D.; Barkun, A. Gastroenterological and hepatic manifestations of patients with COVID-19, prevalence, mortality by country, and intensive care admission rate: Systematic review and meta-analysis. BMJ Open Gastroenterol. 2021, 8, e000571.
- Bolia, R.; Dhanesh Goel, A.; Badkur, M.; Jain, V. Gastrointestinal Manifestations of Pediatric Coronavirus Disease and Their Relationship with a Severe Clinical Course: A Systematic Review and Meta-analysis. J. Trop. Pediatr. 2021, 67, fmab051.
- Russo, T.; Pizuorno, A.; Oskrochi, G.; Latella, G.; Massironi, S.; Schettino, M.; Aghemo, A.; Pugliese, N.; Brim, H.; Ashktorab, H. Gastrointestinal Manifestations, Clinical Characteristics and Outcomes of COVID-19 in Adult and Pediatric Patients. SOJ Microbiol. Infect. Dis. 2021, 8, 109.
- Wang, Y.; Li, Y.; Zhang, Y.; Liu, Y.; Liu, Y. Are gastrointestinal symptoms associated with higher risk of Mortality in COVID-19 patients? A systematic review and meta-analysis. BMC Gastroenterol. 2022, 22, 106.
- Hayashi, Y.; Wagatsuma, K.; Nojima, M.; Yamakawa, T.; Ichimiya, T.; Yokoyama, Y.; Kazama, T.; Hirayama, D.; Nakas, H. The characteristics of gastrointestinal symptoms in patients with severe COVID-19: A systematic review and meta-analysis. J. Gastroenterol. 2021, 56, 409–420.
- Yusuf, F.; Fahriani, M.; Mamada, S.S.; Frediansyah, A.; Abubakar, A.; Maghfirah, D.; Fajar, J.K.; Maliga, H.A.; Ilmawan, M.; Emran, T.B.; et al. Global prevalence of prolonged gastrointestinal symptoms in COVID-19 survivors and potential pathogenesis: A systematic review and meta-analysis. F1000Research 2021, 10, 301.
- Marasco, G.; Maida, M.; Morreale, G.C.; Licata, M.; Renzulli, M.; Cremon, C.; Stanghellini, V.; Barbara, G. Gastrointestinal Bleeding in COVID-19 Patients: A Systematic Review with Meta-Analysis. Can. J. Gastroenterol. Hepatol. 2021, 2021, 2534975.
- Vanella, G.; Capurso, G.; Burti, C.; Fanti, L.; Ricciardiello, L.; Souza Lino, A.; Boskoski, I.; Bronswijk, M.; Tyberg, A.; Krishna Kumar Nair, G.; et al. Gastrointestinal mucosal damage in patients with COVID-19 undergoing endoscopy: An international multicentre study. BMJ Open Gastroenterol. 2021, 8, e000578.
- van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567.
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833.
- Xing, Y.H.; Ni, W.; Wu, Q.; Li, W.J.; Li, G.J.; Wang, W.D.; Tong, J.N.; Song, X.F.; Wing-Kin Wong, G.; Xing, Q.S. Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. J. Microbiol. Immunol. Infect. 2020, 53, 473–480.
- Wu, Y.; Guo, C.; Tang, L.; Hong, Z.; Zhou, J.; Dong, X.; Yin, H.; Xiao, Q.; Tang, Y.; Qu, X.; et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020, 5, 434–435.
- Chen, Y.; Chen, L.; Deng, Q.; Wu, K.; Ni, L.; Yang, Y.; Liu, B.; Wang, W.; Wei, C.; Yang, J.; et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020, 92, 833–840.
- Kang, M.; Wei, J.; Yuan, J.; Guo, J.; Zhang, Y.; Hang, J.; Qu, Y.; Qian, H.; Zhuang, Y.; Chen, X.; et al. Probable Evidence of Fecal Aerosol Transmission of SARS-CoV-2 in a High-Rise Building. Ann. Intern. Med. 2020, 173, 974–980.
- Qian, Q.; Fan, L.; Liu, W.; Li, J.; Yue, J.; Wang, M.; Ke, X.; Yin, Y.; Chen, Q.; Jiang, C. Direct evidence of active SARS-CoV-2 replication in the intestine. Clin. Infect. Dis. 2021, 73, 361–366.
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C. Virological assessment of hospitalized patients with COVID-19. Nature 2020, 581, 465–469.
- Xu, Y.; Li, X.; Zhu, B.; Liang, H.; Fang, C.; Gong, Y.; Guo, Q.; Sun, X.; Zhao, D.; Shen, J.; et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020, 26, 502–505.
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q.; Xie, G.; Lin, S.; Wang, R.; Yang, X.; et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January–March 2020: Retrospective cohort study. BMJ 2020, 369, m1443.
- Jones, D.L.; Baluja, M.Q.; Graham, D.W.; Corbishley, A.; McDonald, J.E.; Malham, S.K.; Hillary, L.S.; Connor, T.R.; Gaze, W.H.; Moura, I.B.; et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020, 749, 141364.
- Han, M.S.; Seong, M.W.; Heo, E.Y.; Park, J.H.; Kim, N.; Shin, S.; Cho, S.I.; Park, S.S.; Choi, E.H. Sequential analysis of viral load in a neonate and her mother infected with SARS-CoV-2. Clin. Infect. Dis. 2020, 71, 2236–2239.
- Pan, Y.; Zhang, D.; Yang, P.; Poon, L.M.; Wang, Q. Viral load of SARSCoV-2 in clinical samples. Lancet Infect. Dis. 2020, 20, 411–412.
- Walsh, K.A.; Jordan, K.; Clyne, B.; Rohde, D.; Drummond, L.; Byrne, P.; Ahern, S.; Carty, P.G.; O’Brien, K.K.; O’Murchu, E.; et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J. Infect. 2020, 81, 357–371.
- Xiao, F.; Sun, J.; Xu, Y.; Li, F.; Huang, X.; Li, H.; Zhao, J.; Huang, J.; Zhao, J. Infectious SARS-CoV-2 in Feces of Patient with Severe COVID-19. Emerg. Infect. Dis. 2020, 26, 1920–1922.
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844.
- Natarajan, A.; Han, A.; Zlitni, S.; Brooks, E.F.; Vance, S.E.; Wolfe, M.; Singh, U.; Jagannathan, P.; Pinsky, B.A.; Boehm, A.; et al. Standardized preservation, extraction and quantification techniques for detection of fecal SARS-CoV-2 RNA. Nat. Commun. 2021, 12, 5753.
- Miyakawa, K.; Machida, M.; Kawasaki, T.; Nishi, M.; Akutsu, H.; Ryo, A. Reduced replication efficacy of severe acute respiratory syndrome coronavirus 2 omicron variant in “mini-gut” organoids. Gastroenterology 2022, 163, 514–516.
- Cheung, K.S.; Hung, I.F.N.; Chan, P.P.Y.; Lung, K.C.; Tso, E.; Liu, R.; Ng, Y.Y.; Chu, M.Y.; Chung, T.W.H.; Tam, A.R.; et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterology 2020, 159, 81–95.
- von Stillfried, S.; Villwock, S.; Bülow, R.D.; Djudjaj, S.; Buhl, E.M.; Maurer, A.; Ortiz-Brüchle, N.; Celec, P.; Klinkhammer, B.M.; Wong, D.W.L.; et al. SARS-CoV-2 RNA screening in routine pathology specimens. Microb. Biotechnol. 2021, 14, 1627–1641.
- Livanos, A.E.; Jha, D.; Cossarini, F.; Gonzalez-Reiche, A.S.; Tokuyama, M.; Aydillo, T.; Parigi, T.L.; Ladinsky, M.S.; Ramos, I.; Dunleavy, K.; et al. Intestinal Host Response to SARS-CoV-2 Infection and COVID-19 Outcomes in Patients with Gastrointestinal Symptoms. Gastroenterology 2021, 160, 2435–2450.
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.G.; van Schayck, J.P.; Mykytyn, A.Z.; Duimel, H.Q.; et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369, 50–54.
- Yang, D.; Perbtani, Y.B.; Loeb, J.; Liu, N.; Draganov, P.V.; Estores, D.E.; Lauzardo, M.; Maurelli, A.; Lednicky, J.A.; Morris, J.G. Detection of SARS-CoV-2 in the gastrointestinal tract among patients with negative nasopharyngeal COVID-19 testing prior to endoscopy. Endosc. Int. Open 2021, 9, E1276–E1282.
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2015; Volume 1282, pp. 1–23.
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574.
- Guo, Y.R.; Cao, Q.D.; Hong, Z.S.; Tan, Y.Y.; Chen, S.D.; Jin, H.J.; Tan, K.S.; Wang, D.Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak. An update on the status. Mil. Med. Res. 2020, 7, 11.
- Arya, R.; Kumari, S.; Pandey, B.; Mistry, H.; Bihani, S.C.; Das, A.; Prashar, V.; Gupta, G.D.; Panicker, L.; Kumar, M. Structural insights into SARS-CoV-2 proteins. J. Mol. Biol. 2021, 433, 166725.
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, e69.
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165878.
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263.
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6.
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734.
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620.
- Zang, R.; Gomez Castro, M.F.; McCune, B.T.; Zeng, Q.; Rothlauf, P.W.; Sonnek, N.M.; Liu, Z.; Brulois, K.F.; Wang, X.; Greenberg, H.B.; et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020, 5, eabc3582.
- Hirose, R.; Nakaya, T.; Naito, Y.; Daidoji, T.; Watanabe, Y.; Yasuda, H.; Konishi, H.; Itoh, Y. Mechanism of human influenza virus RNA persistence and virion survival in feces: Mucus protects virions from acid and digestive juices. J. Infect. Dis. 2017, 216, 105–109.
- Holmes, K.V. Enteric infections with coronaviruses and toroviruses. In Novartis Foundation Symposium; John Wiley: Chichester, UK, 2001.
- Zhou, J.; Li, C.; Zhao, G.; Chu, H.; Wang, D.; Yan, H.H.; Poon, V.K.; Wen, L.; Wong, B.H.; Zhao, X.; et al. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci. Adv. 2017, 3, eaao4966.
- Simsek, C.; Erul, E.; Balaban, H.Y. Role of gastrointestinal system on transmission and pathogenesis of SARS-CoV-2. World J. Clin. Cases 2021, 9, 5427–5434.
- Zhang, M.; Feng, C.; Zhang, X.; Hu, S.; Zhang, Y.; Min, M.; Liu, B.; Ying, X.; Liu, Y. Susceptibility Factors of Stomach for SARS-CoV-2 and Treatment Implication of Mucosal Protective Agent in COVID-19. Front. Med. 2021, 7, 597967.
- Sugimoto, M.; Yamaoka, Y.; Shirai, N.; Furuta, T. Role of renin-angiotensin system in gastric oncogenesis. J. Gastroenterol. Hepatol. 2012, 27, 442–451.
- Balamtekin, N.; Artuk, C.; Arslan, M.; Gülşen, M. The Effect of Helicobacter pylori on the Presentation and Clinical Course of Coronavirus Disease 2019 Infection. J. Pediatr. Gastroenterol. Nutr. 2021, 72, 511–513.
- Lee, S.W.; Ha, E.K.; Yeniova, A.Ö.; Moon, S.Y.; Kim, S.Y.; Koh, H.Y.; Yang, J.M.; Jeong, S.J.; Moon, S.J.; Cho, J.Y.; et al. Severe clinical outcomes of COVID-19 associated with proton pump inhibitors: A nationwide cohort study with propensity score matching. Gut 2021, 70, 76–84.
- Zippi, M.; Fiorino, S.; Budriesi, R.; Micucci, M.; Corazza, I.; Pica, R.; de Biase, D.; Gallo, C.G.; Hong, W. Paradoxical relationship between proton pump inhibitors and COVID-19: A systematic review and meta-analysis. World J. Clin. Cases 2021, 9, 2763–2777.
- Israelsen, S.B.; Ernst, M.T.; Lundh, A.; Lundbo, L.F.; Sandholdt, H.; Hallas, J.; Benfield, T. Proton Pump Inhibitor Use Is Not Strongly Associated With SARS-CoV-2 Related Outcomes: A Nationwide Study and Meta-analysis. Clin. Gastroenterol. Hepatol. 2021, 19, 1845–1854.
- Hamming, I.; Timens, W.; Bulthuis, M.; Lely, A.T.; Navis, G.J.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS Coronavirus. J. Pathol. 2004, 203, 631–637.
- Qi, F.; Qian, S.; Zhang, S.; Zhang, Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020, 526, 135–140.
- Zou, X.; Chen, K.; Zou, J.; Han, P.; Hao, J.; Han, Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020, 14, 185–192.
- Vuille-dit-Bille, R.N.; Camargo, S.M.; Emmenegger, L.; Sasse, T.; Kummer, E.; Jando, J.; Hamie, Q.M.; Meier, C.F.; Hunziker, S.; Forras-Kaufmann, Z.; et al. Human intestine luminal ACE2 and amino acid transporter expression increased by ACE-inhibitors. Amino Acids 2015, 47, 693–705.
- Sungnak, W.; Huang, N.; Bécavin, C.; Berg, M. HCA Lung Biological Network. SARS-CoV-2 Entry Genes Are Most Highly Expressed in Nasal Goblet and Ciliated Cells within Human Airways. arXiv 2020, arXiv:2003.06122v1.
- Zhang, H.; Li, H.B.; Lyu, J.R.; Lei, X.M.; Li, W.; Wu, G.; Lyu, J.; Dai, Z.M. Specific ACE2 expression in small intestinal enterocytes may cause gastrointestinal symptoms and injury after 2019-nCoV infection. Int. J. Infect. Dis. 2020, 96, 19–24.
- Camargo, S.M.; Singer, D.; Makrides, V.; Huggel, K.; Pos, K.M.; Wagner, C.A.; Kuba, K.; Danilczyk, U.; Skovby, F.; Kleta, R.; et al. Tissue-specific amino acid transporter partners ACE2 and collectrin differentially interact with hartnup mutations. Gastroenterology 2009, 136, 872–882.
- Gao, N.; Dou, X.; Yin, T.; Yang, Y.; Yan, D.; Ma, Z.; Bi, C.; Shan, A. Tryptophan Promotes Intestinal Immune Defense through Calcium-Sensing Receptor (CaSR)-Dependent Metabolic Pathways. J. Agric. Food Chem. 2021, 69, 13460–13473.
- Qi, J.; Zhou, Y.; Hua, J.; Zhang, L.; Bian, J.; Liu, B.; Zhao, Z.; Jin, S. The scRNA-seq Expression Profiling of the Receptor ACE2 and the Cellular Protease TMPRSS2 Reveals Human Organs Susceptible to SARS-CoV-2 Infection. Int. J. Environ. Res. Public Health 2021, 18, 284.
- Pearce, S.C.; Suntornsaratoon, P.; Kishida, K.; Al-Jawadi, A.; Guardia, J.; Nadler, I.; Flores, J.; Shiarella, R.; Auvinen, M.; Yu, S.; et al. Expression of SARS-CoV-2 entry factors, electrolyte, and mineral transporters in different mouse intestinal epithelial cell types. Physiol. Rep. 2021, 9, e15061.
- Singh, M.; Bansal, V.; Feschotte, C. A Single-Cell RNA Expression Map of Human Coronavirus Entry Factors. Cell Rep. 2020, 32, 108175.
- Zhou, L.; Niu, Z.; Jiang, X.; Zhang, Z.; Zheng, Y.; Wang, Z.; Zhu, Y.; Gao, L.; Huang, H.; Wang, X.; et al. SARS-CoV-2 Targets by the pscRNA Profiling of ACE2, TMPRSS2 and Furin Proteases. iScience 2020, 23, 101744.
- Johnson, B.A.; Xie, X.; Bailey, A.L.; Kalveram, B.; Lokugamage, K.G.; Muruato, A.; Zou, J.; Zhang, X.; Juelich, T.; Smith, J.K.; et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 2021, 591, 293–299.
- Papa, G.; Mallery, D.L.; Albecka, A.; Welch, L.G.; Cattin-Ortolá, J.; Luptak, J.; Paul, D.; McMahon, H.T.; Goodfellow, I.G.; Carteret, A.; et al. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathog. 2021, 17, e1009246.
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865.
- Berni Canani, R.; Comegna, M.; Paparo, L.; Cernera, G.; Bruno, C.; Strisciuglio, C.; Zollo, I.; Gravina, A.G.; Miele, E.; Cantone, E.; et al. Age-Related Differences in the Expression of Most Relevant Mediators of SARS-CoV-2 Infection in Human Respiratory and Gastrointestinal Tract. Front. Pediatr. 2021, 9, 697390.
- Zhang, F.; Li, W.; Feng, J.; Ramos da Silva, S.; Ju, E.; Zhang, H.; Chang, Y.; Moore, P.S.; Guo, H.; Gao, S.J. SARS-CoV-2 pseudovirus infectivity and expression of viral entry-related factors ACE2, TMPRSS2, Kim-1, and NRP-1 in human cells from the respiratory, urinary, digestive, reproductive, and immune systems. J. Med. Virol. 2021, 93, 6671–6685.
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860.
- Helal, M.A.; Shouman, S.; Abdelwaly, A.; Elmehrath, A.O.; Essawy, M.; Sayed, S.M.; Saleh, A.H.; El-Badri, N. Molecular basis of the potential interaction of SARS-CoV-2 spike protein to CD147 in COVID-19 associated-lymphopenia. J. Biomol. Struct. Dyn. 2022, 40, 1109–1119.
- Fenizia, C.; Galbiati, S.; Vanetti, C.; Vago, R.; Clerici, M.; Tacchetti, C.; Daniele, T. SARS-CoV-2 entry: At the crossroads of CD147 and ACE2. Cells 2021, 10, 1434.
- Shilts, J.; Crozier, T.W.M.; Greenwood, E.J.D.; Lehner, P.J.; Wright, G.J. No evidence for basigin/CD147 as a direct SARS-CoV-2 spike binding receptor. Sci. Rep. 2021, 11, 413.
- Bortolotti, D.; Simioni, C.; Neri, L.M.; Rizzo, R.; Semprini, C.M.; Occhionorelli, S.; Laface, I.; Sanz, J.M.; Schiuma, G.; Rizzo, S.; et al. Relevance of VEGF and CD147 in different SARS-CoV-2 positive digestive tracts characterized by thrombotic damage. FASEB J. 2021, 35, e21969.
- Wang, H.; Ye, J.; Liu, R.; Chen, G.; Zhao, J.; Huang, L.; Yang, F.; Li, M.; Zhang, S.; Xie, J.; et al. Clinical significance of CD147 in children with inflammatory bowel disease. Biomed. Res. Int. 2020, 2020, 7647181.
- Tamhane, T.; Arampatzidou, M.; Gerganova, V.; Tacke, M.; Illukkumbura, R.; Dauth, S.; Schaschke, N.; Peters, C.; Reinheckel, T.; Brix, K. The activity and localization patterns of cathepsins B and X in cells of the mouse gastrointestinal tract differ along its length. Biol. Chem. 2014, 395, 1201–1219.
- Tamhane, T.; Lllukkumbura, R.; Lu, S.; Maelandsmo, G.M.; Haugen, M.H.; Brix, K. Nuclear cathepsin L activity is required for cell cycle progression of colorectal carcinoma cells. Biochimie 2016, 122, 208–218.
This entry is offline, you can click here to edit this entry!
