Please note this is an old version of this entry, which may differ significantly from the current revision.
Foodborne diseases caused by contaminated food, including foodborne pathogens, seriously threaten public health and the economy. This has led to the development of more sensitive and accurate methods for detecting pathogenic bacteria. Hybridization chain reaction (HCR), an isothermal nucleic acid hybridization signal amplification technique, has received increasing attention due to its enzyme-free and isothermal characteristics, and pathogenic bacteria detection methods using HCR for signal amplification have experienced rapid development in the last five years.
- hybridization chain reaction
- foodborne pathogens
- food safety
1. Introduction
With the continuous development of international food trade and the globalization of the food market, food safety is still worthy of extensive attention. As we all know, foodborne pathogens (bacteria, fungi, and viruses) widely exist in human daily life. They quickly cause foodborne diseases by contaminating food in food production, processing, storage, and transportation, which poses a significant threat to public health, personal health, and the national economy [1][2]. According to the World Health Organization (WHO), foodborne diseases cause 600 million cases and 420,000 deaths worldwide each year, of which 30% of foodborne deaths occur, especially in 5-year-old children, and the loss of 33 million disability-adjusted life years (DALY) [3][4]. The most common foodborne pathogens include Salmonella. Enteritidis (S. enteritidis), Salmonella Typhimurium (S. Typhimurium), Campylobacter, Staphylococcus aureus (S. aureus), Listeria monocytogenes (L. monocytogenes), and Escherichia coli O157:H7 (E. coli O157:H7) [5][6][7]. It is worth noting that some bacteria and fungi produce toxins during their growth and reproduction, and even many of these pathogens and toxins are thermostable, and therefore cannot be easily destroyed by typical food preparation methods (cooking, frying, freezing, etc.) [8]. Early detection of foodborne pathogens is necessary to prevent outbreaks of foodborne diseases, improve overall cost-effectiveness, and ensure better food safety management [9].
For the detection of pathogenic bacteria, the traditional culture method needs to go through the procedures of pathogenic bacteria cultivation, isolation, recovery, and identification, which often takes 3–7 days and has the disadvantages of requiring professional staff, being time-consuming, and having a complicated operation process, as well as being a false negative and lacking sensitivity, which is challenging to adapt to the development of rapid, accurate, and sensitive detection technology. With the development of immunology and molecular biology, enzyme-linked immunosorbent assays (ELISA) and polymerase chain reaction (PCR) have become the gold standard for detecting foodborne pathogens. However, both methods have their drawbacks. ELISA, which is based on the immune response, is limited by the complexity of antibody preparation and lot-to-lot variations. Based on nucleic acid amplification, PCR has high sensitivity but requires specialized personnel and expensive equipment and is prone to cross-contamination and false positives [10][11][12][13]. The development of timely, sensitive, accurate, and cost-effective technologies for detecting foodborne pathogens has therefore been a challenging task in food safety. In order to achieve the sensitivity required to detect pathogens at deficient concentrations in complex food matrices, signal amplification techniques such as enzyme-linked cascade amplification and nucleic acid amplification [14] are increasingly developing and applied for pathogen detection to meet the growing demand for food safety.
Some bacteria (such as emetic Bacillus cereus) are non-antigenic and lack antibodies. In addition, proteins themselves cannot be amplified, such as antibodies or antigens. Therefore, nucleic acid-based signal amplification has more significant potential advantages and generally falls into two categories: thermal cycle amplification techniques, such as PCR and ligase chain reaction (LCR) [15]; and isothermal amplification techniques, including rolling circle amplification (RCA), loop-mediated isothermal amplification (LAMP), strand displacement amplification (SDA), hybridization chain reaction (HCR), recombinase polymerase amplification (RPA), etc. Isothermal amplification is an emerging nucleic acid amplification method attractive to many food production industries due to its constant temperature, cost-effectiveness, and the absence of expensive thermal cycling equipment in thermal cycle amplification techniques.
2. HCR in Foodborne Pathogen Detection
The ideal pathogen detection method should be rapid, sensitive, specific, economical, equipment-free, and intuitive. Therefore, developing new detection methods that meet the comprehensive performance requirements of quantification, sensitivity, specificity, and economy is urgently needed. Among them, HCR has attracted much attention due to its advantages, such as being enzyme-free, having isothermal conditions, its self-assembly, and excellent amplification efficiency. In addition, new technology for pathogen detection often needs to have three parts: identification, transduction or amplification, and output signal. Among them, HCR is based on the specific recognition and triggering characteristics of target nucleic acid and has both easy modification of signal molecule transduction (such as electrochemical reagents [16], nanoparticles [17], fluorescent dyes [18], biotin, etc.) and good amplification ability, so that it can easily combined with various signal output platforms (such as colorimetry, fluorescence, electrochemistry, Raman scattering, chemiluminescence, etc.), which has excellent potential for further expansion in the field of foodborne pathogen detection [19][20][21][22].
2.1. Colorimetry
Colorimetry is a typical biochemical assay that detects analytes by comparing or measuring the color change caused by a specific reaction between the analyte and the sensing material, either by the naked eye or by a simple optical instrument, which allows for intuitive qualitative or quantitative detection [23][24][25]. The colorimetric method is widely used because of its simplicity, rapidity, cost-effectiveness, and visualization of results [26]. Colorimetric results usually include changes in a single-color shade and color type. The first type is represented by enzymes (e.g., natural enzymes, nano enzymes, DNA enzymes), which cause changes in single color intensity due to the difference in the catalytic ability of the enzyme on the chromogenic substrate; the second type is represented by nanoparticles (gold and silver nanoparticles, nanoclusters), which cause a change in color through changes in extinction coefficient caused by aggregation, growth, etching, etc. However, the sensitivity of colorimetric sensing systems is sometimes unsatisfactory [27]. Considering improvements by introducing technical solutions such as HCR signal amplification to achieve sensitive detection is more than welcome.
In 2016, the Lai group first reported a sandwich ELISA for detecting Escherichia coli O157:H7 (E. coli O157:H7) in milk using HCR loaded with horseradish peroxidase (HRP) [28]. A gold nanoparticle probe (mAb-AuNP-DNA1) modified with specific antibodies and trigger chains was used as a signal transduction and amplification carrier. In the presence of E. coli O157:H7, a “pAb/E. coli O157:H7/mAb-AuNP-DNA1” sandwich complex was formed on the ELISA substrate, followed by the addition of biotin-modified hairpin probes (H1-biotin, H2-biotin). Triggered by DNA1, HCR generated long-nicked dsDNA rich in biotin sites. After the addition of horseradish peroxidase (HRP-SA) anchored with streptavidin and chromogenic substrate (TMB + H2O2), this sandwich ELISA method had a detection limit of 1.08 × 102 CFU/mL in the concentration range of 5 × 102–1 × 107 CFU/mL of E. coli O157:H7, which was 185 times lower than that of the traditional ELISA (Figure 1A).
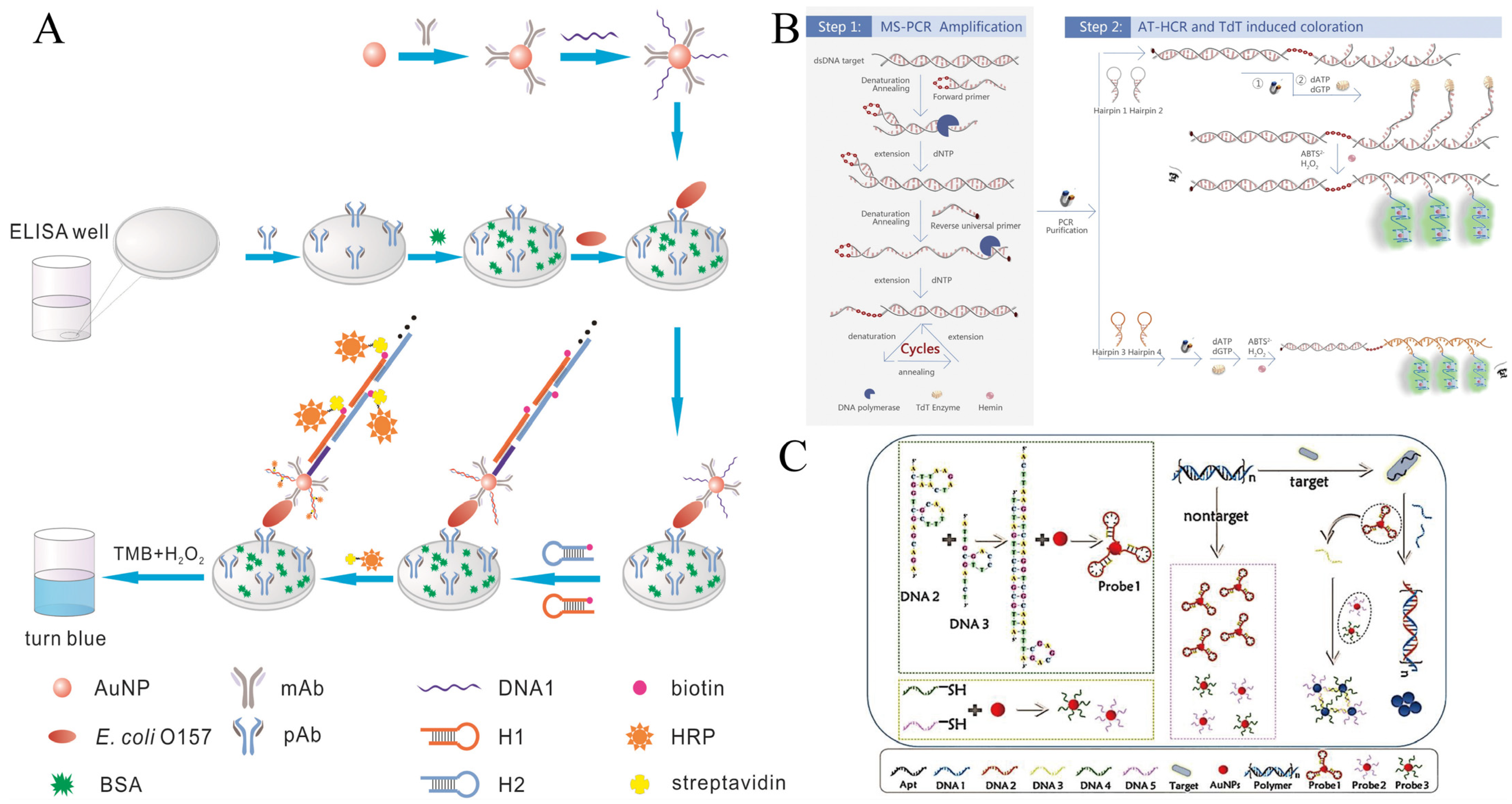
Figure 1. HCR-based colorimetric sensing platform for foodborne pathogen detection. (A) Sandwich type of ELISA using HCR combined with natural enzymes. (B) Dual amplification using PCR-HCR combined with GQH DNAzyme. (C) AuNP-based colorimetric sensing combined with HCR strategies works mainly through two mechanisms.
Furthermore, not only can nucleases (DNAzyme) solve the unstable and expensive problems of natural enzymes and the editable properties of nucleic acids with the compatible rivalry of HCR’s editable nature, but HCR-DNAzyme can be developed into a universal paradigm. Tian et al. proposed an ultrasensitive colorimetric dual-target detection platform based on HCR-G-quadruplex/hemin DNAzyme (HCR-GQH DNAzyme) [29]. First, this method allows for multiplex super PCR (MS-PCR) simultaneous dual-target amplification—the reverse primer labeled biotin for binding with magnetic probes to eliminate primer dimer interference. In contrast, the forward primer is blocked by inserting an ethylene glycol bridge inhibitor to limit the extension of the polymerase, resulting in a large number of dsDNA products with single-strand DNA (ssDNA) toes protruding (ssDNA-dsDNA). Then, two pairs of hairpin probes containing 5 nt oligonucleotide tails (H1, H2, and H3, H4) perform their respective HCR reactions, producing many HCR products containing 5 nt oligonucleotide tails. The interference hairpin is removed by magnetic separation, and Terminal deoxynucleotidyl Transferase (TdT) catalyzed 5-nt ssDNA tail extension to amplify, then form GQH DNAzyme, which catalyzes the oxidation of ABTS to produce a green color. This visual detection method creatively combines MS-PCR and AT-HCR-DNAzyme to simultaneously detect Salmonella and Staphylococcus aureus in fat-free milk samples with a 10 CFU/mL detection limit (Figure 1B).
In addition, AuNP is also popular for its property of changing the color of the solution due to aggregation and dispersion. However, the sensitivity of sensors based only on unlabeled aptamers and AuNP has been reported to reach only 105 CFU/mL [30]. Therefore, it is urgent to improve the detection sensitivity of such colorimetric methods by increasing the reaction efficiency, cycling amplification [31][32], or introducing signal amplification technologies such as HCR.
Sun et al. innovatively combined two mechanisms to develop a highly sensitive dual-signal amplification colorimetric strategy [33] based on DNA@AuNPs (Probe 1 and Probe 2/3) aggregation, in which Probe 1 is formed by the interaction between AuNPs and sticky-ended short ssDNA, and Probe 2/3 utilizes thiolated DNA to stabilize AuNPs via “Au-S” bonds (Figure 1C). When the target is present, the polymer releases DNA 1, which activates the HCR, and thus induces the aggregation of Probe 1. Meanwhile, DNA 3, as a linker, separates from Probe 1 and further assembles Probe 2/3. The more sensitive color response of the system is due to the synergistic aggregation of two different DNA@AuNPs probes triggered by one activation. Under optimal conditions, the ultrasensitive AuNPs-based strategy achieved sensitive detection of 10 CFU/mL and was successfully applied to detect E. coli from tap water and milk samples.
However, single-color colorimetric methods continue to face the problem of being difficult to distinguish and having low color resolution. With the development of nanoscience, various AuNP-derived structures have emerged (such as gold rods, gold nanobipyramids, gold nanostars, silver nanoprisms, etc.) that break the shackles of a single color. Due to their sharp morphology, small changes at hot spots easily lead to sensitive changes in extinction coefficients, resulting in rainbow-like color changes, promoting the broader development of colorimetric methods.
Tang et al. reported a high-color-resolution and ultrasensitive detection biosensor [34] for the detection of methicillin-resistant Staphylococcus aureus (mecA gene) in milk based on the multi-HCR of AuNPs and the alkaline phosphatase (ALP)-mediated in situ growth of gold nanobipyramid particles (AuNBPs) (Figure 2A). In the presence of target DNA, it acts as a linking bridge to hybridize magnetic capture probes (CP) and AuNP signal probes (SP), carrying plenty of ALP. With the assistance of NADPH, a series of rainbow-colored results of the in situ growth of AuNBPs with different concentrations of target DNA are observed. This method can detect as low as 2.71 pM target DNA through multi-step amplification of AuNP vectors, multi-HCR amplification, and enzymatic reactions. Importantly, the multiple color output results are easy to determine visually.
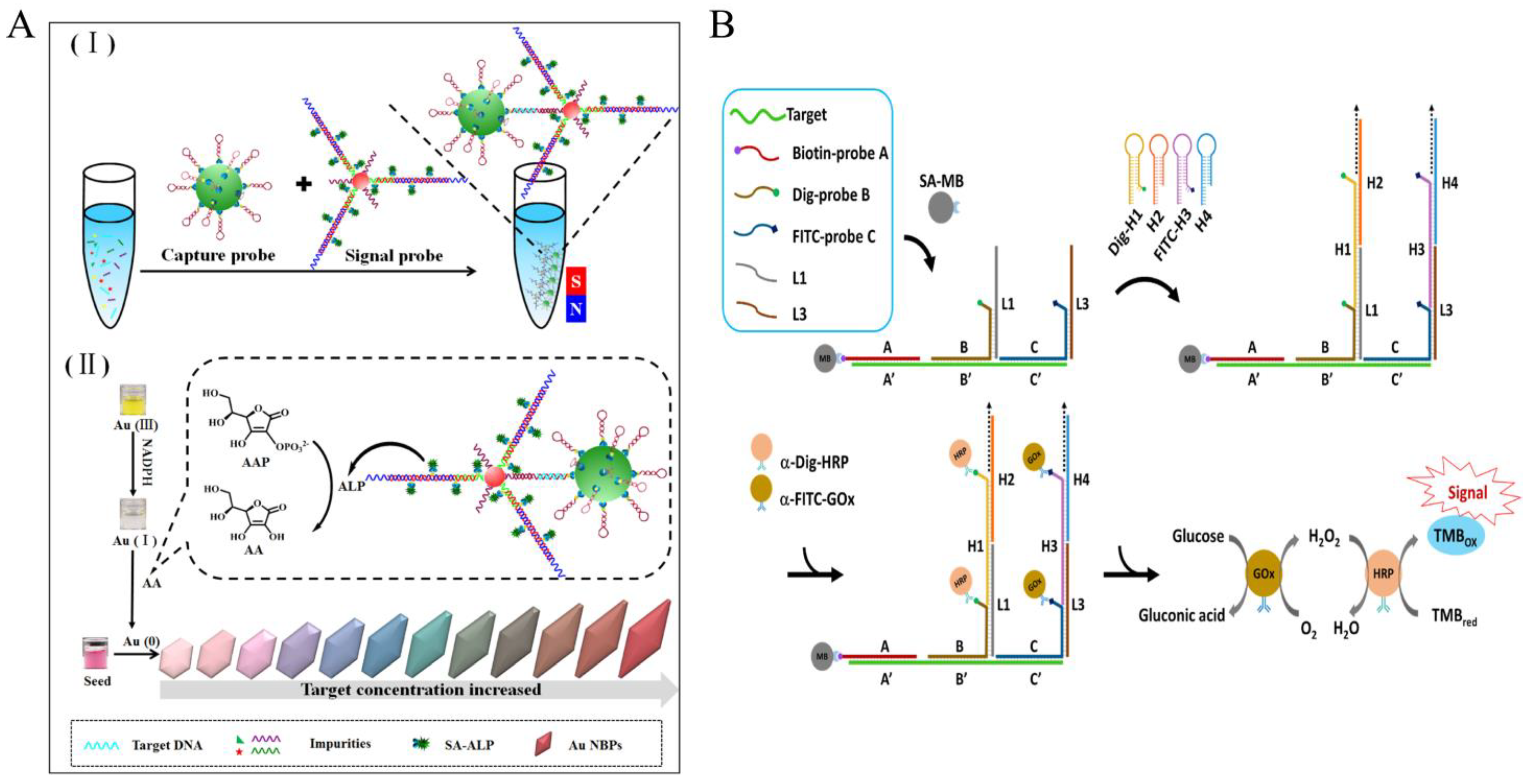
Figure 2. HCR-based colorimetric sensing platform for foodborne pathogen detection. (A) Multi-color colorimetric strategy using multi-HCR combined with gold nanobipyramids. (I) the hybridization process of capture probe, target DNA and signal probe; (II) the signal producing process by using NADPH-assisted ALP-mediated in situ growth of AuNBPs. (B) DNA logic-gate circuits using HCR combined with enzymatic cascade amplification.
At the same time, the powerful editable capability of HCR not only significantly enhances specificity but also enables cascade amplification of the enzyme. Yang et al. by using a DNA logic gate strategy and combining Dig-Probe and FITC-probe strategies (Figure 2B). Only when three specific regions of the L. monocytogenes specific genomic sequences are present simultaneously is the HCR reaction triggered to form a long dsDNA that is enriched with glucose oxidase (GOx) and is HRP-rich. GOx converts glucose to gluconate and produces hydrogen peroxide, allowing it to further mediate the oxidation of TMB by HRP, producing a highly specific visual color signal output for enzyme cascade amplification with a detection limit of 1.12 nM [35], and successfully applied to a variety of food matrices such as vegetables, meat, and milk extracts.
2.2. Fluorescence
Fluorescence (FL)-based methods have a higher sensitivity compared to colorimetric methods, with fluorescence spectra as the resultant response. The current fluorescence strategies using HCR for pathogen detection are broadly divided into two types [36]: (1) labeling of fluorescent groups or insertion of fluorescent dyes; (2) fluorescence energy transfer, including fluorescence resonance energy transfer (FRET) and contact quenching (CQ) [37].
The first type is more cost-effective than the others and is commonly used, but the sensitivity is somewhat reduced by the use of a single fluorescent group or dye with its high background fluorescence interference. Therefore, it is often necessary to separate or adsorb the background fluorescence with the aid of various nanomaterials (e.g., magnetic beads, graphene oxide, MnO2, MXene,) to obtain an accurate and sensitive fluorescence signal.
Yu et al. proposed a fluorescent sensor for detecting Salmonella (S. Enteritidis, S. Typhimurium, and S. Choleraesuis) invA gene in lettuce based on HCR using MBs as a separator [38]. Long ssDNA prepared by asymmetric polymerase chain reaction (aPCR) was subsequently captured by capture probe-modified MBs, the exposed target ssDNA triggered HCR amplification of FAM-labelled reporter probes (H1-FAM, H2-FAM), and finally, magnetic separation and boiling released the reacted hairpin reporter probes, achieving a limit of detection of 7.4 × 101 CFU/mL in buffer and 6.9 × 102 CFU/g in spiked lettuce (Figure 3A). There is also some research using a combined strategy of magnetic beads and HCR, resulting in low-cost, reusable recycling [39] and high-throughput detection [40].
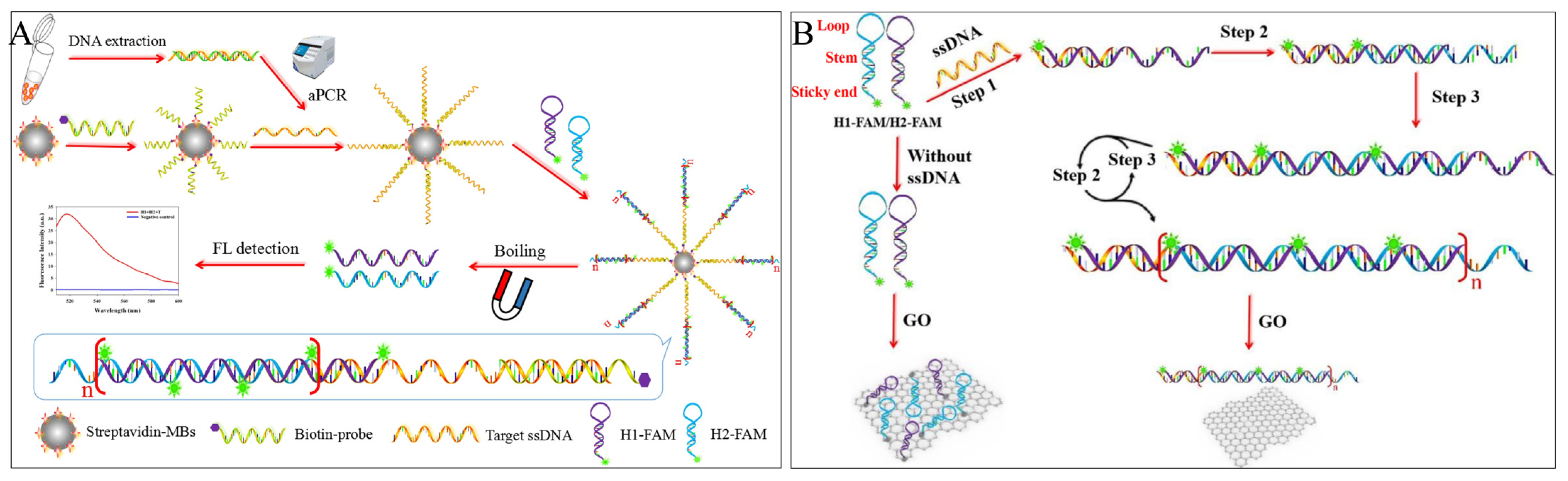
Figure 3. HCR-based fluorescence sensing platform using the carrier for foodborne pathogen detection. (A) A direct method used with magnetic beads. (B) A “HCR-GO” platform.
Apart from MBs, the two-dimensional nanomaterial—graphene oxide (GO) [41] is commonly preferred for its excellent fluorescence quenching properties for most fluorescent dyes, fluorescent groups, quantum dots, and metal fluorescent nanoclusters. In addition, it has been cleverly used as an adjunct to fluorescence strategies in HCR technology due to its different affinity for ssDNA and dsDNA and as an “HCR-GO” platform to reduce background values or restore fluorescence signals. Yu et al. proposed a fluorescence “turn-on” strategy based on the “HCR-GO” platform [42]. Initially, the hairpin single strands were quenched by GO, and fluorescence was turned off. When Salmonella ssDNA was present, HCR was triggered by the alternating hybridization of H1-FAM and H2-FAM to form a long dsDNA nanowire, which detached from the GO surface and restored fluorescence. The detection of Salmonella ssDNA was completed within 3.5 h, with a detection limit of 4.2 × 101 CFU/mL in buffer and 4.2 × 102 CFU/mL in milk (Figure 3B).
With the aid of nanotechnology, various HCR-based assisted co-amplification strategies (e.g., multi-technology co-amplification, enzyme-assisted HCR, fluorescent synergy effect, etc.) have been proposed. Liang et al. coupled variable temperature amplification with isothermal amplification techniques to establish a novel PCR-HCR dual-signal amplification fluorescence method [43], effectively increasing the sensitivity of the assay to 100 times that of PCR signal amplification alone, with a detection limit of 7.2 × 101 CFU/mL in buffer and 7.2 × 102 CFU/mL in milk (Figure 4A). Zhang et al. coupled two isothermal amplification techniques to synergistically amplify the detection signal and developed a DNA fluorescence sensor combining HCR and three-way cleavage-assisted signal amplification (3WJ-NEASA) for dual-signal amplification for the first time [44]. The target DNA of S. aureus initiates self-assembly between H1 and H2, exposing the gap to capture molecular beacon to form the 3WJ structure, which is continuously cycled by Nt.BbvcI cleavage enzymes to cleave and capture the molecular beacon, resulting in the accumulation of recovered fluorescence. The benign cycle of the HCR circuit and the 3WJ-NEASA circuit amplifies synergistically, allowing the biosensor to detect S. aureus DNA down to 6.7 pM in one step within 30 min. Finally, the HCR-mediated 3WJ-NEASA method detects S. aureus with a low LOD of 1.2 × 101 CFU/mL and 1.3 × 102 CFU/mL in milk (Figure 4B).

Figure 4. HCR-based fluorescence sensing platform using various auxiliary collaborative amplification strategies for foodborne pathogen detection. (A) Multi-technology amplification. (B) Enzyme-assisted HCR. (C) Synergistic effect.
In addition, the HCR technique can easily combine dsDNA dyes (e.g., SYBR Green I) and FAM fluorophores to produce a fluorescence synergy effect to enhance the fluorescence signal due to the generation of double-stranded products and ease of modification. Tang et al. combined the “FAM-SYBR Green I” fluorescent synergy effect with the “HCR-GO” platform and achieved sensitive detection of S. aureus 16S rRNA with a LOD of 50 pM (Figure 5C). It was also successfully applied to detect milk samples with a LOD of 4 × 102 CFU/mL [45]. The Lu group proposed two ultrasensitive fluorescent detection strategies for methicillin-resistant S. aureus mRNA based on the synergistic effect of “FAM-SYBR Green I” on the “HCR-GO” platform and further incorporating other cyclic amplification strategies [46][47].
FRET can effectively reduce its background fluorescence by bringing the donor-acceptor fluorophores closer together without the aid of additional nanomaterials, which can be easily achieved by HCR self-assembly. Ren et al. proposed a simple FRET-based fluorescence sensor for the detection of Vibrio parahaemolyticus (V. parahaemolyticus) ssDNA using a four-way migration HCR [48]. When ssDNA is present, the auxiliary strand (R) first binds to the target strand to form short dsDNA, which triggers the four-way migration of HCR, and FRET occurs. The method is capable of detecting 0.067 nM ssDNA (Figure 5A). In addition to the use of HCR self-assembly to close the distance between fluorescent groups, FRET-sensitive V. parahaemolyticus detection strategies have also been investigated based on the formation of dsDNA by triple oligonucleotide (TFO) insertion [49].
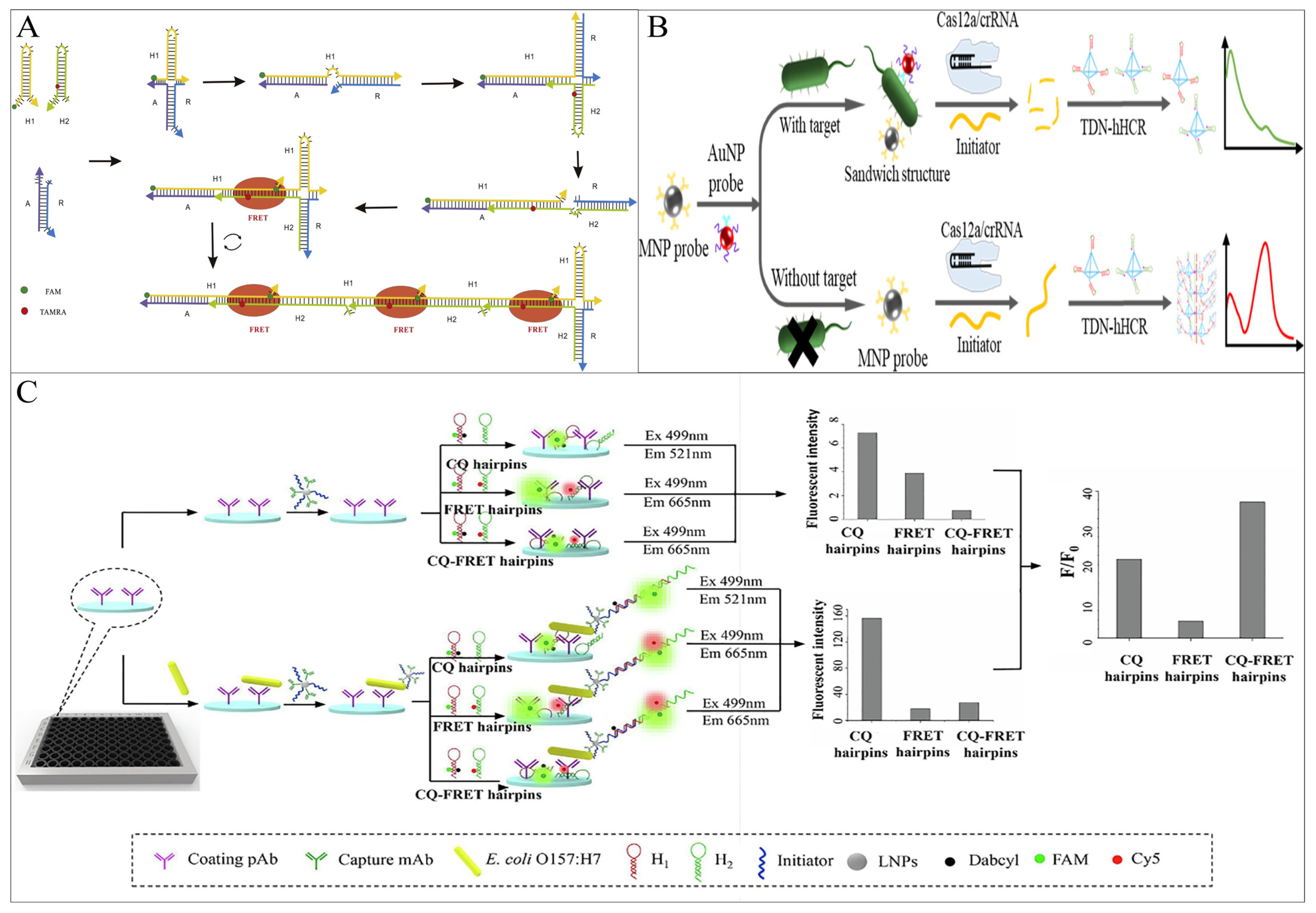
Figure 5. HCR-based fluorescence sensing platform using energy transfer strategies for foodborne pathogen detection. (A) A simple FRET-based method using a four-way migration HCR. (B) A complex FRET-based method using tetrahedral DNA nanostructure-mediated HCR combined with CRISPR-Cas12a. (C) An ELISA method combining FRET and CQ.
The ratio fluorescence output is another feature that makes FRET more popular, thus greatly reducing the interference of uncontrollable factors such as light scattering and photobleaching and obtaining more accurate results. Cai et al. [50] used the “IMB/target/AuNP” sandwich structure to achieve bacterial cell signal converted and amplified into a biological barcode DNA molecule, which triggers the trans-cleavage activity of CRISPR-Cas12a, cleaving the initiator chain of TDN-hHCR, resulting in the failure of the HCR product formation and no FRET occurring between hairpin fluorophores. In contrast, in the absence of Salmonella, CRISPR-Cas12a cannot be activated, and the initiator is intact, triggering the TDN-hHCR process and promoting FRET. This method has been applied to actual samples, achieving Salmonella sensitive detection of 17 CFU/mL in milk and 25 CFU/mL in egg white (Figure 5B).
CQ is another fundamental mechanism of fluorescence energy transfer. When the fluorescent group and the quencher are in proximity in this system, the energy of the fluorescent group can be transferred to the quencher through proton-coupled electron transfer, leading to fluorescence contact quenching [51]. As the distance between them increases, the fluorescence on the fluorescent group can be restored, resulting in a fluorescent signal. The Lai group coupled CQ and ratio FRET through HCR technology, proposing a sandwich ELISA method for the detection of E. coli O157:H7 in milk. They designed a low-background HCR hairpin probe (CQ-FRET probe) to investigate a more sensitive and accurate FRET output strategy [52]. The detection limit of this CQ-FRET method was 3.5 × 101 CFU/mL, significantly lower than that of the CQ hairpin-based immune HCR (3.28 × 103 CFU/mL) and the FRET hairpin-based immune HCR (6.49 × 104 CFU/mL), which were 93-fold and 1854-fold higher, respectively. But there is no doubt that it also increases costs and expenses (Figure 5C).
Finally, it is noteworthy that most of the current HCR-based fluorescence sensing strategies are downconverted fluorescence strategies using high-energy ultraviolet as the excitation light source, which have some obvious drawbacks such as photobleaching, background auto-fluorescence, and cell-damaging properties, limiting their real-time applications. Near-infrared (NIR)-excited upconversion fluorescence strategies represented by lanthanide-doped upconversion nanoparticles (UCNPs) are receiving much attention in the fields of cell imaging and tumor therapy [53]. Recently, the upconversion strategy has also been applied to HCR-based fluorescence detection of foodborne pathogens [54], where the HCR technique amplifies the strongly stabilized fluorescence signal, in addition to the NIR excitation mode, which also has the potential to penetrate deeply into food monitoring.
2.3. Electrochemistry
Electrochemical biosensors typically employ three main kinds of signals as their readout, namely current, voltage, and impedance signal, to reflect the concentrations of analytes [55][56][57][58]. Due to its excellent signal amplification and easy labeling, HCR can load with catalytic elements (e.g., natural enzymes [59], nanoenzymes [60], etc.) or conductive substances (e.g., redox molecules [61], conductive nanomaterials [62], etc.) through modification, electrostatic adsorption, or embedding to influence the electrical signal transduction capacity of the electrode [63][64]. Electrochemical approaches with HCR as the signal amplifier have already attracted more and more attention in bioassays due to their excellent sensitivity as low as the attomolar level [65].
A sensitive electrochemical strategy for the detection of S. aureus in milk and pear juice based on DB-HCR was proposed by Wu et al (Figure 6A). The use of DB-HCR to produce dense products is a perfect solution to the problem of loss of sensitivity in electrochemistry that is often caused by the production of long linear products that deviate from the electrode in conventional HCR. The presence of a target leads to the release of the T-strand from the “Apt-T” complex and its capture by the electrode surface capture probe, triggering the DB-HCR on the electrode surface to generate a dense negatively charged DNA product that attracts a bulk loading of modified positively charged poly methylene blue nanoparticles (pMBNPs), which achieves ultrasensitive detection down to 1 CFU/mL and demonstrates the excellent amplification potential of DB-HCR in electrochemical sensors [66].
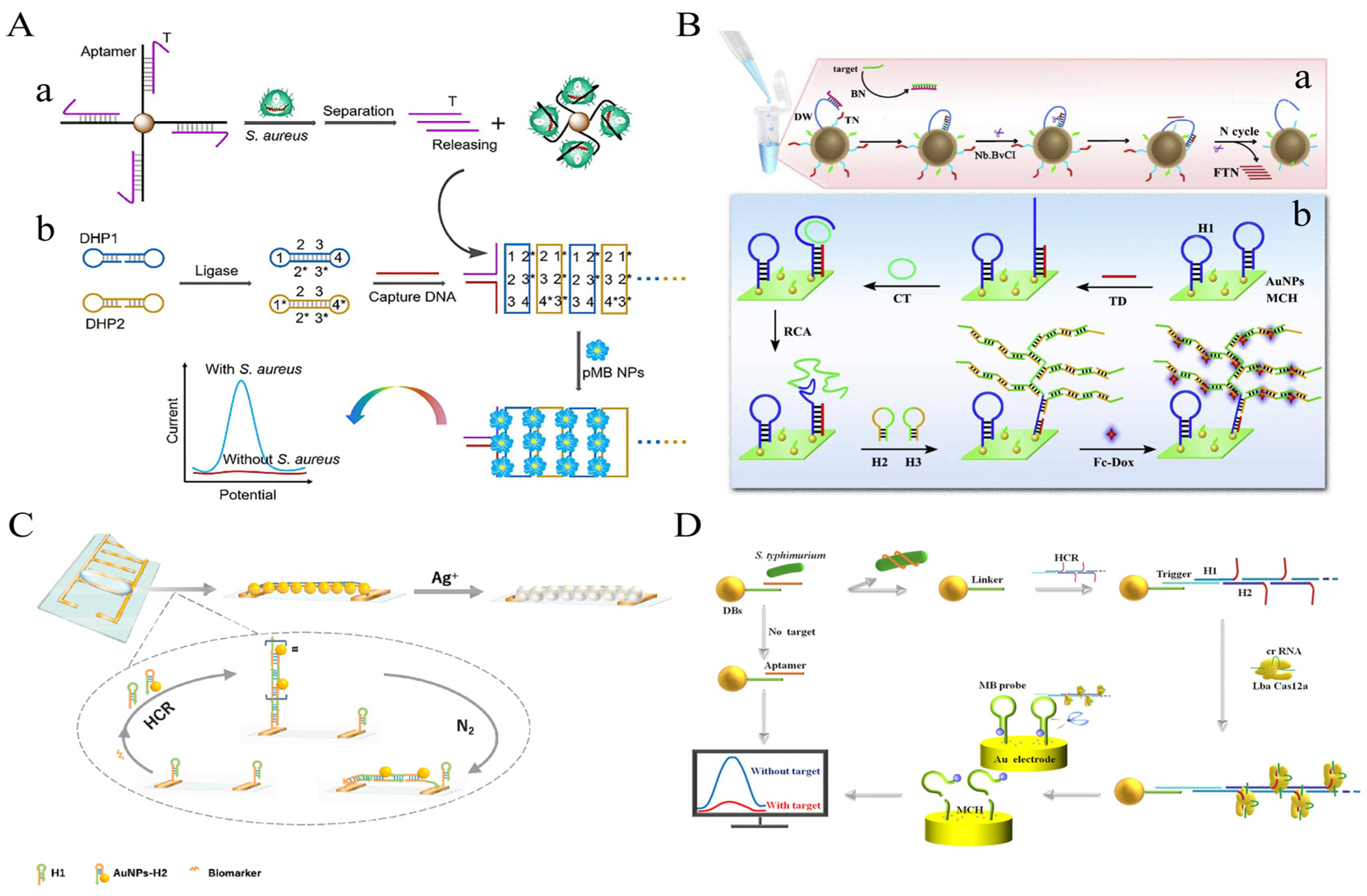
Figure 6. HCR-based electrochemical sensing platform for foodborne pathogen detection. (A) Dumbbell HCR combined with poly methylene blue nanoparticles. (B) Dendritic HCR combined with Fc-Dox. (a) the schematic diagram of the 3D DNA walker-based amplification reactions triggered by the target gene to produce many fragment of TN (FTN); (b) the schematic diagram of HCR and RCA-based amplification reactions on the electrode surface to generate to long double-stranded DNA sequences to immobilize many electrochemical indicators related with the concentration of target gene. (AuNPs, gold nanoparticles; BN, Blocking DNA; CT, Circular template; DW, DNA walker; FTN, fragment of TN; H1, hairpin DNA 1; H2, hairpin DNA 2; H3, hairpin DNA 3; HCR, hybridization chain reaction; MCH, 6-mercapto ethanol; RCA, rolling circle amplification; TN, transfer oligonucleotide) (C) A facile synthesis of silver wire on the electrode combined with AuNPs. (D) The reporter probe on the surface of the electrode was cleaved by HCR-CRISPR/Cas12a system.
Li et al. proposed an ultrasensitive electrochemical biosensor based on 3D DNA Walker, RCA, and HCR multiplex amplification [67]. By generating dendritic DNA products by HCR, the spatial site resistance problem, often present in electrochemical receptors, is avoided. This assay demonstrated high sensitivity to the target DNA, with a 7 CFU/mL LOD (Figure 6B).
In addition, Feng et al. cleverly exploited the characteristics of HCR-grown linear DNA nanowires to propose a simple and novel electrochemical scheme of simple synthetic silver wires between electrodes to detect S. aureus 16S rRNA in milk [68]. In the presence of 16S rRNA, the modified sub-stable hairpin probe H1 on the electrode surface opened, activating HCR. The stem-loop structures of H1 and the hairpin probe H2 modified with AuNPs (H2-AuNPs) alternately opened to finally form a long dsDNA-RNA (HCR product)-AuNPs product. Under the influence of N2 cycles, the modified HCR product of the AuNPs was tiled in the electrode gap to form a framework, and silver was deposited on the AuNP of the extended ladder product between the electrodes to form silver wires, leading to a drastic change in electrical parameters. This method achieved a detection limit of 50 CFU/mL within 50–107 CFU/mL of S. aureus, with a total time of 100 min (Figure 6C).
In recent years, the clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas) systems have been widely reported in the field of biosensing due to their high-turnover non-specific endonuclease activity [69][70]. However, the sensitivity of the CRISPR/Cas system only reaches the pM level, and it is often necessary to use pre-signal amplification methods or engineered circuits to extend the detection limit [71][72]. Among them, the HCR-CRISPR strategy has been used to detect various targets. Cas12a can recognize dsDNA and initiate trans-cleavage activity under the guidance of crRNA but requires a pro-spacer adjacent motif (PAM) sequence adjacent to the target dsDNA, which may limit the broad application of its HCR. To address these issues, Liu et al. introduced the PAM sequence into the amplification product of HCR to construct a sensitive and rapid electrochemical assay for the detection of S. Typhimurium in milk [73]. In the presence of S. Typhimurium, Aptamer on IMB preferentially binds to the target. The naked Linker strand binds to the trigger strand of the HCR product that forms many ssDNA side chains, which activates the trans-cleavage activity of the Cas12a-crRNA complex without the PAM sequence, thereby cleaving the surface-modified electrical signal response of the Au electrode ssDNA reporter probes. When the target is absent, no trans cleavage activity of Cas12a is observed, thereby retaining the reporter probe on the surface of the Au electrode. The method allows for the selective and sensitive quantification of S. Typhimurium with a 20 CFU/mL LOD (Figure 6D).
To address the criticism of electrochemistry’s inherent instability and poor reproducibility [74], Wang et al. proposed a fluorescent-electrochemical dual-signal sensing strategy based on magnetic DNA-Walker and HCR coupled amplification for the detection of Clostridium perfringens (C. perfringens) [75]. The method can significantly improve accuracy and detect C. perfringens at a minimum concentration of 1 CFU/g in meat products (e.g., chicken, beef, duck, mutton, and pork) through a multi-modal signal output approach.
This entry is adapted from the peer-reviewed paper 10.3390/foods12224067
References
- Bintsis, T. Foodborne Pathogens. AIMS Microbiol. 2017, 3, 529–563.
- Dewey-Mattia, D. Surveillance for Foodborne Disease Outbreaks—United States, 2009–2015. MMWR Surveill. Summ. 2018, 67, 1.
- Food Safety. Available online: https://www.who.int/news-room/fact-sheets/detail/food-safety (accessed on 13 August 2023).
- Estimating the Burden of Foodborne Diseases. Available online: https://www.who.int/activities/estimating-the-burden-of-foodborne-diseases (accessed on 13 August 2023).
- Foodborne Pathogens. Available online: http://www.aimspress.com/article/10.3934/microbiol.2017.3.529 (accessed on 13 August 2023).
- Food Science of Animal Resources. Available online: https://www.kosfaj.org/archive/view_article?pid=kosfa-41-1-1 (accessed on 13 August 2023).
- Most Common Foodborne Pathogens and Mycotoxins on Fresh Produce: A Review of Recent Outbreaks: Critical Reviews in Food Science and Nutrition. Available online: https://www.tandfonline.com/doi/abs/10.1080/10408398.2013.777021?journalCode=bfsn20 (accessed on 13 August 2023).
- Martinović, T.; Andjelković, U.; Gajdošik, M.Š.; Rešetar, D.; Josić, D. Foodborne Pathogens and Their Toxins. J. Proteom. 2016, 147, 226–235.
- Weng, X.; Zhang, C.; Jiang, H. Advances in Microfluidic Nanobiosensors for the Detection of Foodborne Pathogens. LWT 2021, 151, 112172.
- Sugiarti, S.A.; Nurhayati, N. Optimization of Annealing Temperature for Detection of Lipase Gene in Bacillus subtilis Using Polymerase Chain Reaction (PCR) Method. J. Phys. Conf. Ser. 2021, 1725, 012046.
- Nurmasytha, A.; Yuliati, F.N.; Hajrawati; Prahesti, K.I. Microbiological Analysis of Raw Chicken Meat Sold at Maros Traditional Markets: Total Plate Count and Escherichia coli. IOP Conf. Ser. Earth Environ. Sci. 2021, 788, 012118.
- You, S.-M.; Luo, K.; Jung, J.-Y.; Jeong, K.-B.; Lee, E.-S.; Oh, M.-H.; Kim, Y.-R. Gold Nanoparticle-Coated Starch Magnetic Beads for the Separation, Concentration, and SERS-Based Detection of E. coli O157:H7. ACS Appl. Mater. Interfaces 2020, 12, 18292–18300.
- Zeng, L.; Guo, L.; Wang, Z.; Xu, X.; Ding, H.; Song, S.; Xu, L.; Kuang, H.; Xu, C. Gold Nanoparticle-Based Immunochromatographic Assay for Detection Pseudomonas Aeruginosa in Water and Food Samples. Food Chem. X 2021, 9, 100117.
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545.
- Ding, Y.; Choo, J.; deMello, A.J. From Single-Molecule Detection to next-Generation Sequencing: Microfluidic Droplets for High-Throughput Nucleic Acid Analysis. Microfluid. Nanofluid. 2017, 21, 58.
- Sheikhzadeh, E.; Beni, V.; Zourob, M. Nanomaterial Application in Bio/Sensors for the Detection of Infectious Diseases. Talanta 2021, 230, 122026.
- Guo, Y.; Zhou, Y.; Fu, J.; Fang, H.; Li, Y.; Huang, X.; Xiong, Y. A Self-Luminous Bifunctional Bacteria Directed Fluorescent Immunosensor for the Simultaneous Detection and Quantification of Three Pathogens in Milk. Sens. Actuators B Chem. 2021, 338, 129757.
- Zhang, H.; Mou, J.; Ding, J.; Qin, W. Magneto-Controlled Potentiometric Assay for E. coli Based on Cleavage of Peptide by Outer-Membrane Protease T. Electrochim. Acta 2021, 384, 138408.
- Yang, X.; Yu, Y.; Gao, Z. A Highly Sensitive Plasmonic DNA Assay Based on Triangular Silver Nanoprism Etching. ACS Nano 2014, 8, 4902–4907.
- Dai, S.; Xue, Q.; Zhu, J.; Ding, Y.; Jiang, W.; Wang, L. An Ultrasensitive Fluorescence Assay for Protein Detection by Hybridization Chain Reaction-Based DNA Nanotags. Biosens. Bioelectron. 2014, 51, 421–425.
- Chen, L.; Sha, L.; Qiu, Y.; Wang, G.; Jiang, H.; Zhang, X. An Amplified Electrochemical Aptasensor Based on Hybridization Chain Reactions and Catalysis of Silver Nanoclusters. Nanoscale 2015, 7, 3300–3308.
- Li, N.; Chen, J.; Luo, M.; Chen, C.; Ji, X.; He, Z. Highly Sensitive Chemiluminescence Biosensor for Protein Detection Based on the Functionalized Magnetic Microparticles and the Hybridization Chain Reaction. Biosens. Bioelectron. 2017, 87, 325–331.
- Colorimetric Biosensor Based on Smartphone: State-of-Art—ScienceDirect. Available online: https://www.sciencedirect.com/science/article/pii/S0924424722006914 (accessed on 14 August 2023).
- Xu, N.; Jin, S.; Wang, L. Metal Nanoparticles-Based Nanoplatforms for Colorimetric Sensing: A Review. Rev. Anal. Chem. 2020, 40, 1–11.
- Light-Controlled Configurable Colorimetric Sensing Array. Analytical Chemistry. Available online: https://pubs.acs.org/doi/full/10.1021/acs.analchem.9b00549 (accessed on 14 August 2023).
- Yang, W.; Weng, C.; Li, X.; He, H.; Fei, J.; Xu, W.; Yan, X.; Zhu, W.; Zhang, H.; Zhou, X. A Sensitive Colorimetric Sensor Based on One-Pot Preparation of h-Fe3O4@ppy with High Peroxidase-like Activity for Determination of Glutathione and H2O2. Sens. Actuators B Chem. 2021, 338, 129844.
- Wu, J.; Lv, J.; Zheng, X.; Wu, Z.-S. Hybridization Chain Reaction and Its Applications in Biosensing. Talanta 2021, 234, 122637.
- Guo, Q.; Han, J.-J.; Shan, S.; Liu, D.-F.; Wu, S.-S.; Xiong, Y.-H.; Lai, W.-H. DNA-Based Hybridization Chain Reaction and Biotin–Streptavidin Signal Amplification for Sensitive Detection of Escherichia coli O157:H7 through ELISA. Biosens. Bioelectron. 2016, 86, 990–995.
- Tian, J.; Huang, K.; Luo, Y.; Zhu, L.; Xu, Y.; Xu, W. Visual Single Cell Detection of Dual-Pathogens Based on Multiplex Super PCR (MS-PCR) and Asymmetric Tailing HCR (AT-HCR). Sens. Actuators B Chem. 2018, 260, 870–876.
- Wu, W.; Li, M.; Wang, Y.; Ouyang, H.; Wang, L.; Li, C.; Cao, Y.; Meng, Q.; Lu, J. Aptasensors for Rapid Detection of Escherichia coli O157:H7 and Salmonella Typhimurium. Nanoscale Res. Lett. 2012, 7, 658.
- Yang, H.; Xiao, M.; Lai, W.; Wan, Y.; Li, L.; Pei, H. Stochastic DNA Dual-Walkers for Ultrafast Colorimetric Bacteria Detection. Anal. Chem. 2020, 92, 4990–4995.
- Song, S.; Wang, X.; Xu, K.; Xia, G.; Yang, X. Visualized Detection of Vibrio Parahaemolyticus in Food Samples Using Dual-Functional Aptamers and Cut-Assisted Rolling Circle Amplification. J. Agric. Food Chem. 2019, 67, 1244–1253.
- Liu, M.; Geng, L.; Zhang, F.; Dou, S.; Li, F.; Liu, Z.; Guo, Y.; Sun, X. Isolation of Bacteria Aptamers with Non-SELEX for the Development of a Highly Sensitive Colorimetric Assay Based on Dual Signal Amplification. J. Agric. Food Chem. 2022, 70, 15990–15998.
- Zhou, J.; Fu, R.; Liu, H.; Liu, Y.; Wang, Y.; Jiao, B.; He, Y.; Tang, H. Integrating Multiple Hybridization Chain Reactions on Gold Nanoparticle and Alkaline Phosphatase-Mediated in Situ Growth of Gold Nanobipyramids: An Ultrasensitive and High Color Resolution Colorimetric Method to Detect the mecA Gene of Staphylococcus aureus. J. Hazard. Mater. 2021, 418, 126223.
- Yang, F.-A.; Wu, Y.-T.; Liu, Y.-W.; Liao, W.-C. Hybridization Chain Reaction-Assisted Enzyme Cascade Genosensor for the Detection of Listeria monocytogenes. Talanta 2023, 254, 124193.
- Zhou, R.; Zeng, Z.; Sun, R.; Liu, W.; Zhu, Q.; Zhang, X.; Chen, C. Traditional and New Applications of the HCR in Biosensing and Biomedicine. Analyst 2021, 146, 7087–7103.
- Zeng, Z.; Zhou, R.; Sun, R.; Zhang, X.; Cheng, Z.; Chen, C.; Zhu, Q. Nonlinear Hybridization Chain Reaction-Based Functional DNA Nanostructure Assembly for Biosensing, Bioimaging Applications. Biosens. Bioelectron. 2021, 173, 112814.
- Yu, S.; Tang, Y.; Yan, M.; Aguilar, Z.P.; Lai, W.; Xu, H. A Fluorescent Cascade Amplification Method for Sensitive Detection of Salmonella Based on Magnetic Fe3O4 Nanoparticles and Hybridization Chain Reaction. Sens. Actuators B Chem. 2019, 279, 31–37.
- Chu, Z.J.; Xiao, S.J.; Liu, Y.H.; Xiong, G.L.; Huang, D.J.; Wang, S.P.; Zhao, X.J.; Zhang, Z.B. Rapid and Sensitive Detection of the IS6110 Gene Sequences of Mycobacterium tuberculosis Based on Hybridization Chain Reaction and Reusable Magnetic Particles. Sens. Actuators B Chem. 2019, 282, 904–909.
- Yu, B.; Li, F.; Zhao, T.; Li, F.; Zhou, B.; Xu, H. Hybridization Chain Reaction-Based Flow Cytometric Bead Sensor for the Detection of Emetic Bacillus Cereus in Milk. Sens. Actuators B Chem. 2018, 256, 624–631.
- Zhang, Y.; Chen, X.; Roozbahani, G.M.; Guan, X. Graphene Oxide-Based Biosensing Platform for Rapid and Sensitive Detection of HIV-1 Protease. Anal. Bioanal. Chem. 2018, 410, 6177–6185.
- Yu, S.; Xu, Q.; Huang, J.; Yi, B.; Aguilar, Z.P.; Xu, H. Rapid and Sensitive Detection of Salmonella in Milk Based on Hybridization Chain Reaction and Graphene Oxide Fluorescence Platform. J. Dairy Sci. 2021, 104, 12295–12302.
- Liang, T.; Wu, X.; Chen, B.; Liu, J.; Aguilar, Z.P.; Xu, H. The PCR-HCR Dual Signal Amplification Strategy for Ultrasensitive Detection of Escherichia coli O157:H7 in Milk. LWT 2020, 130, 109642.
- Zhang, C.; Luo, Z.; Wu, M.; Ning, W.; Tian, Z.; Duan, Y.; Li, Y. A Highly Sensitive Fluorescence Biosensor for Detection of Staphylococcus aureus Based on HCR-Mediated Three-Way DNA Junction Nicking Enzyme Assisted Signal Amplification. Analyst 2021, 146, 6528–6536.
- Tang, J.; Wang, Z.; Zhou, J.; Lu, Q.; Deng, L. Enzyme-Free Hybridization Chain Reaction-Based Signal Amplification Strategy for the Sensitive Detection of Staphylococcus aureus. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 215, 41–47.
- Ning, Y.; Chen, S.; Hu, J.; Li, L.; Cheng, L.; Lu, F. Fluorometric Determination of agrA Gene Transcription in Methicillin-Resistant Staphylococcus aureus with a Graphene Oxide–Based Assay Using Strand-Displacement Polymerization Recycling and Hybridization Chain Reaction. Microchim. Acta 2020, 187, 372.
- Wang, X.; Liu, S.; Xiao, R.; Hu, J.; Li, L.; Ning, Y.; Lu, F. Graphene-Oxide-Based Bioassay for the Fluorometric Determination of agrC Gene Transcription in Methicillin-Resistant Staphylococcus aureus That Uses Nicking-Enzyme-Assisted Target Recycling and a Hybridization Chain Reaction. Talanta 2022, 250, 123714.
- Ren, D.; Sun, C.; Huang, Z.; Luo, Z.; Zhou, C.; Li, Y. A Novel FRET Biosensor Based on Four-Way Branch Migration HCR for Vibrio Parahaemolyticus Detection. Sens. Actuators B Chem. 2019, 296, 126577.
- Tan, X.-H.; Li, Y.-B.; Liao, Y.; Liu, H.-Z. An Enzyme Free Fluorescence Resonance Transfer Strategy Based on Hybrid Chain Reaction and Triplex DNA for Vibrio Parahaemolyticus. Sci. Rep. 2020, 10, 20710.
- Cai, Q.; Shi, H.; Sun, M.; Ma, N.; Wang, R.; Yang, W.; Qiao, Z. Sensitive Detection of Salmonella Based on CRISPR-Cas12a and the Tetrahedral DNA Nanostructure-Mediated Hyperbranched Hybridization Chain Reaction. J. Agric. Food Chem. 2022, 70, 16382–16389.
- Marras, S.A.E.; Kramer, F.R.; Tyagi, S. Efficiencies of Fluorescence Resonance Energy Transfer and Contact-mediated Quenching in Oligonucleotide Probes. Nucleic Acids Res. 2002, 30, e122.
- Duan, M.; Xiao, X.; Huang, Y.; Li, G.; Shan, S.; Lv, X.; Zhou, H.; Peng, S.; Liu, C.; Liu, D.; et al. Immuno-HCR Based on Contact Quenching and Fluorescence Resonance Energy Transfer for Sensitive and Low Background Detection of Escherichia coli O157:H7. Food Chem. 2021, 334, 127568.
- Ma, C.; Bian, T.; Yang, S.; Liu, C.; Zhang, T.; Yang, J.; Li, Y.; Li, J.; Yang, R.; Tan, W. Fabrication of Versatile Cyclodextrin-Functionalized Upconversion Luminescence Nanoplatform for Biomedical Imaging. Anal. Chem. 2014, 86, 6508–6515.
- Xu, Y.; Ahmad, W.; Hassan, M.M.; Li, H.; Ouyang, Q.; Chen, Q. Ultrasensitive Hairpin Mediated Upconversion Fluorescence Biosensor for Staphylococcus aureus Detection in Foods and Waters Exploiting g–C3N4–Assisted Catalysis. Anal. Chim. Acta 2023, 1239, 340738.
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical Biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763.
- Cho, M.; Lee, S.; Han, S.-Y.; Park, J.-Y.; Rahman, M.A.; Shim, Y.-B.; Ban, C. Electrochemical Detection of Mismatched DNA Using a MutS Probe. Nucleic Acids Res. 2006, 34, e75.
- Flexible and Wearable Electrochemical Biosensors Based on Two-Dimensional Materials: Recent Developments. SpringerLink. Available online: https://link.springer.com/article/10.1007/s00216-020-03002-y?utm_source=xmol&utm_medium=affiliate&utm_content=meta&utm_campaign=DDCN_1_GL01_metadata (accessed on 14 August 2023).
- Frontiers. State-of-Art Bio-Assay Systems and Electrochemical Approaches for Nanotoxicity Assessment. Available online: https://www.frontiersin.org/articles/10.3389/fbioe.2020.00325/full (accessed on 14 August 2023).
- Lee, J.; Tatsumi, A.; Tsukakoshi, K.; Wilson, E.D.; Abe, K.; Sode, K.; Ikebukuro, K. Application of a Glucose Dehydrogenase-Fused with Zinc Finger Protein to Label DNA Aptamers for the Electrochemical Detection of VEGF. Sensors 2020, 20, 3878.
- Two-Dimensional Transition Metal Carbides as Supports for Tuning the Chemistry of Catalytic Nanoparticles. Nature Communications. Available online: https://www.nature.com/articles/s41467-018-07502-5?utm_source=xmol&utm_medium=affiliate&utm_content=meta&utm_campaign=DDCN_1_GL01_metadata (accessed on 14 August 2023).
- Li, H.; Dauphin-Ducharme, P.; Ortega, G.; Plaxco, K.W. Calibration-Free Electrochemical Biosensors Supporting Accurate Molecular Measurements Directly in Undiluted Whole Blood. J. Am. Chem. Soc. 2017, 139, 11207–11213.
- An Enzyme-Free Electrochemical Sandwich DNA Assay Based on the Use of Hybridization Chain Reaction and Gold Nanoparticles: Application to the Determination of the DNA of Helicobacter Pylori. SpringerLink. Available online: https://link.springer.com/article/10.1007/s00604-019-3999-z (accessed on 14 August 2023).
- Yang, H.; Gao, Y.; Wang, S.; Qin, Y.; Xu, L.; Jin, D.; Yang, F.; Zhang, G.-J. In Situ Hybridization Chain Reaction Mediated Ultrasensitive Enzyme-Free and Conjugation-Free Electrochemcial Genosensor for BRCA-1 Gene in Complex Matrices. Biosens. Bioelectron. 2016, 80, 450–455.
- Xiong, E.; Zhang, X.; Liu, Y.; Zhou, J.; Yu, P.; Chen, J. An Electrochemical Biosensor for Sensitive Detection of Hg2+ Based on Exonuclease III-Assisted Target Recycling and Hybridization Chain Reaction Amplification Strategies. Anal. Methods 2016, 8, 2106–2111.
- Zhang, Q. Application of Hybridization Chain Reaction (HCR) in Electrochemical Analysis. Int. J. Electrochem. Sci. 2022, 17, 220227.
- Wu, T.; Wang, C.; Wu, M.; Wang, P.; Feng, Q. Novel Integrating Polymethylene Blue Nanoparticles with Dumbbell Hybridization Chain Reaction for Electrochemical Detection of Pathogenic Bacteria. Food Chem. 2022, 382, 132501.
- Li, Y.; Liu, H.; Huang, H.; Deng, J.; Fang, L.; Luo, J.; Zhang, S.; Huang, J.; Liang, W.; Zheng, J. A Sensitive Electrochemical Strategy via Multiple Amplification Reactions for the Detection of E. coli O157: H7. Biosens. Bioelectron. 2020, 147, 111752.
- Feng, Y.; Zhou, D.; Gao, L.; He, F. Electrochemical Biosensor for Rapid Detection of Bacteria Based on Facile Synthesis of Silver Wire across Electrodes. Biosens. Bioelectron. 2020, 168, 112527.
- Shao, N.; Han, X.; Song, Y.; Zhang, P.; Qin, L. CRISPR-Cas12a Coupled with Platinum Nanoreporter for Visual Quantification of SNVs on a Volumetric Bar-Chart Chip. Anal. Chem. 2019, 91, 12384–12391.
- Zhang, L.; Jiang, H.; Zhu, Z.; Liu, J.; Li, B. Integrating CRISPR/Cas within Isothermal Amplification for Point-of-Care Assay of Nucleic Acid. Talanta 2022, 243, 123388.
- Li, S.-Y.; Cheng, Q.-X.; Wang, J.-M.; Li, X.-Y.; Zhang, Z.-L.; Gao, S.; Cao, R.-B.; Zhao, G.-P.; Wang, J. CRISPR-Cas12a-Assisted Nucleic Acid Detection. Cell Discov. 2018, 4, 20.
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic Acid Detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442.
- Liu, X.; Bu, S.; Feng, J.; Wei, H.; Wang, Z.; Li, X.; Zhou, H.; He, X.; Wan, J. Electrochemical Biosensor for Detecting Pathogenic Bacteria Based on a Hybridization Chain Reaction and CRISPR-Cas12a. Anal. Bioanal. Chem. 2022, 414, 1073–1080.
- Wu, C.; Wu, X.; Hou, F.; Wu, L.; Liu, G. An Ultrasensitive Electrochemical Aptasensor Based on Pd@PCN-222 as a Signal Probe Coupled with Exonuclease III-Assisted Cycling Amplification for the Detection of Ochratoxin A. Food Control 2022, 139, 109066.
- Wang, W.; Yuan, W.; Wang, D.; Mai, X.; Wang, D.; Zhu, Y.; Liu, F.; Sun, Z. Dual-Mode Sensor Based on the Synergy of Magnetic Separation and Functionalized Probes for the Ultrasensitive Detection of Clostridium Perfringens. RSC Adv. 2022, 12, 25744–25752.
This entry is offline, you can click here to edit this entry!
 Encyclopedia
Encyclopedia
