A prevalent, well-characterized immobilization design system is the biotin–(strept)avidin interaction. The biotin–(strept)avidin interaction is considered to be one of the most specific and stable non-covalent interaction, whose dissociation constant (KD) is about 103 to 106 times higher than an antigen–antibody interaction. Its high affinity is principally useful for isolating and amplifying the signal, which increases the ability for the detection of very low concentrations of analyte while decreasing the number of steps required for measurement, allowing for a more rapid quantitation of analyte. The biotin–(strept)avidin system offers enormous advantages over other covalent and non-covalent interactions, which include amplification of weak signals, efficient operation, robustness, and astonishing stability against manipulation, proteolytic enzymes, temperature and pH extremes, harsh organic reagents, and other denaturing reagents. Since the biotin–(strept)avidin interaction is one of the strongest known non-covalent interactions in nature, avidin and its analogues have therefore been extensively used as probes and affinity matrices for a wide variety of applications in the field of biotechnology, such as biochemical assays, diagnostics, affinity purification, and drug delivery.
1. History
The first immunoassay was developed in 1959 by Solomon Berson and Rosalyn S. Yalow for the detection of insulin in patient blood samples [
1]. Their previous work on insulin metabolism helped them recognize that antibodies can be used to study this hormone [
2]. They discovered that the competition of endogenous human insulin with radioactively labeled (I
131) tyrosine residues of bovine insulin in binding to the anti-insulin antibodies in the sera of guinea pigs immunized with bovine insulin [
35] provided a basis for a sensitive and specific assay to quantify insulin in patients [
2]. This first radioimmunoassay won the Nobel Prize in Medicine in 1977 and paved the way for the development of other types of immunoassays [
1]. In 1971, Eva Engvall and Paul Perlmann pioneered the enzyme-linked immunosorbent assay (ELISA) by immobilizing antigens on a solid support to measure the concentration of antibodies in a sample using an enzyme-linked anti-immunoglobulin detection antibody. Simultaneously, B.K. van Weeman and A.H. Schuurs developed an assay with the same principles quantifying an antigen, rather than an antibody [
13]. Since then, the ELISA method has become a standard of routine laboratory research and diagnostic methods worldwide [
36].
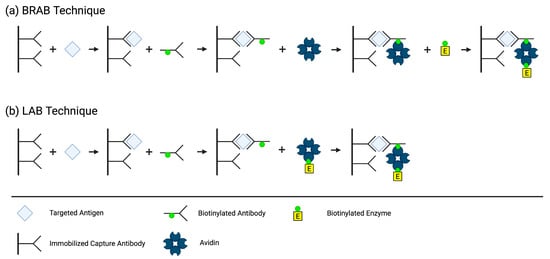
In 1979, Jean-Luc Guesdon, Therese Ternynck, and Stratis Avrameas, a team from the Institut Pasteur in France, developed the first ELISA employing the avidin–biotin system. They created two different methods by conjugating biotin to antibodies, antigens, and enzymes, which then binds to avidin to detect, immobilize, and quantify antibodies and/or antigens of interest. In the Bridged Avidin–Biotin (BRAB,
Figure 3a) method, the antigen from the sample is “sandwiched” between an immobilized capture antibody and a biotin-labeled antibody. After a washing step eliminating the unbound biotin-labeled antibody, avidin is added and binds to the biotin label on the immune complex. A second washing step occurs to eliminate the unbound avidin, followed by the addition of a biotin-labeled enzyme that binds to the immobilized avidin. Lastly, a third washing step removes the unbound biotin-labeled enzyme, and the enzyme activity associated with the presence of antigen in the sample is measured. In the Labeled Avidin–Biotin (LAB,
Figure 3b) technique, the antigen from the sample is bound to an immobilized antibody and a biotin-labeled antibody, just like in the BRAB technique. However, in the LAB technique, the avidin is pre-labeled with the enzyme, eliminating the need for an extra step [
37].
Figure 3. Methods of BRAB (a) and LAB (b). Created using BioRender.com accessed 29 October 2023.
The applications and designs of the biotin–streptavidin system are vast, giving researchers the ability to perform a variety of tasks including identification, localization, and quantitation of the target analyte. The system allows for an indirect interaction between two biomolecules, preserving the natural binding properties of the antibodies and antigens. Biotin has the ability to be conjugated to a wide variety of biomolecules without altering the interaction of the biomolecule with its target ligand [
38]. Biotin’s relatively small size (240 Da), flexible valeric side chain [
39], and ease of conjugation [
27] make it well suited to protein labeling [
40].
2. Biotin
Biotin was identified from a seemingly simple observation; consuming raw egg whites leads to toxicity, and this toxicity can be prevented using an unknown substance in the egg yolk [
41]. In 1916, W. G. Bateman of Yale University made the casual observation that raw egg whites in the diet of animals had a toxic effect. Then, 11 years later, Margaret A. Boas at the Lister Institute of Preventative Medicine in London came across the same phenomenon. By feeding raw egg whites to rats as the source of protein in their diet, she observed that they developed dermatitis and hemorrhages of the skin, loss of hair, paralyzed limbs, lost considerable weight, and eventually died. Cooking the egg whites did not elicit toxic symptoms and its effects could be alleviated or prevented by the consumption of food containing biotin. Egg whites contain the toxic protein avidin, which is neutralized via its binding to biotin (a component of egg yolks), preventing its absorption and toxic effects. Biotin was then discovered in 1936 by Fritz Kögl and B. Tönnis from the University of Utrecht, Netherlands, who were looking to find a source of a minute quantity of an unknown compound present in a charcoal fraction of “brewer’s wort” (an extract of ground malt) that was necessary for the growth of yeast cultures. Kögl found that egg yolks contained the highest concentration he could find of this substance. In a series of 16 tedious steps from 550 pounds of dried duck egg yolks, Kögl and B. Tönnis succeeded in isolating the active principle of this fraction, which ultimately turned out to be biotin [
42].
Biotin (vitamin H, B
7 [
43], coenzyme R [
29]) is a water-soluble B-complex vitamin and essential coenzyme for five human carboxylases (acetyl–CoA carboxylase (ACC) 1 & 2, pyruvate carboxylase (PC), propionyl–CoA carboxylase (PCC), and 3-methylcrotonyl–CoA carboxylase (MCC)) [
44]. It is composed of a tetrahydrothiophene ring, a valeric acid side chain, and a ureido (tetrahydroimidizalone) ring [
45] that functions as the carrier for CO
2 [
41]. Biotin has a well-studied role as a covalently bound coenzyme that catalyzes critical steps in the metabolic pathways of gluconeogenesis, fatty acid synthesis and oxidation, and amino acid catabolism. But more recently, it has been revealed that biotin has noncarboxylic functions involved in cell signaling, the epigenetic regulation of genes and the chromatin structure, and immune response [
44]. Biotin binds to avidin with a K
D in the order of ~10
−15 M and binds to streptavidin with a K
D in the order of ~10
−14 M [
29], forming the strongest known noncovalent ligand–protein interaction, achieving picomolar and even femtomolar affinities [
39]. Binding occurs very rapidly and remains undisturbed by pH and temperature extremes, organic solvents, and other denaturing agents, making this a stable and ideal system to use in immunoassays [
29].
3. (Strept)avidin
Avidins are a family of biotin-binding proteins that are found in both eukaryotic and prokaryotic species. The first known avidin was isolated from chicken (
Gallus gallus) egg whites in 1941 [
46] by researchers Robert E. Eakin, Esmond E. Snell, and Roger J. Williams. They confirmed that the constituent in raw egg whites capable of inactivation using biotin in vitro [
47] and eliciting toxic effects when fed to animals [
48] is a protein called avidin [
49], named for its avidity with biotin (
avidity + biot
in) [
50]. The first bacterial avidin, streptavidin, was isolated from the antibiotic-secreting
Streptomyces avidinii bacteria in 1964. Since then, several new avidins have been experimentally verified from both eukaryotic and prokaryotic species [
46]. Several genetically and chemically engineered avidins and their analogues have also been studied to advance our knowledge about the functional and structural characteristics of avidins, potentially leading to more successful applications [
27].
Avidin and streptavidin are functional and structural [
51] tetrameric glycoprotein [
27] analogues that contain four biotin-binding sites [
51]. Although the functional properties and the quaternary and tertiary structures of these two proteins are well conserved, their primary structures have low similarity (~30%) [
46]. Each of the four monomers contains eight antiparallel β-strands that form a β-barrel. At one end of each β-barrel is a biotin-binding site that comprises several aromatic residues [
39] and a tryptophan (Trp) residue from the neighboring subunit. The residues from the biotin-binding site bind with the ureido group of biotin using hydrogen bonds and van der Waals interactions [
52]. A flexible loop of 12 and 8 amino acids of avidin and streptavidin, respectively, closes the binding pocket on top of biotin, which contributes significantly to its binding strength [
5]. The tryptophan from the adjacent monomer acts as a hydrophobic lid for the binding pocket [
45], making it one of the most important amino acid residues responsible for the tight biotin interaction [
53]. Two of the monomers lie parallel to each other, forming a dimer with an extensive interface, and the two dimers associate, forming a weaker interface. Interestingly, the weaker dimeric interface appears to be important to the high affinity as mutations either increase or decrease the protein’s affinity to biotin [
5]. A mutation in Trp-110 and Trp-120 in avidin and streptavidin, respectively [
45], results in only monomers, which significantly reduces the affinity (K
D~10
−7 M). As new applications of (strept)avidin–biotin complexes are explored, more avidin derivatives will surely continue to emerge [
5].
Chivers et al. engineered an avidin derivative called traptavidin, which exhibits stronger biotin-binding due to two mutations [
39] that stabilize the binding pocket loop by reducing its flexibility [
54] and reducing its conformational change upon biotin binding [
55]. Additionally, researchers have engineered other avidin-derived proteins in attempt to circumvent the pitfalls of avidin [
34], such as a reduced binding affinity when biotin is conjugated to a protein [
29], non-specific binding, and immunogenicity [
27]. Although avidin has a stronger biotin-binding affinity, its basic isoelectric point (pI ~10) makes it prone to nonspecific adsorption of negatively charged molecules [
56] and surfaces such as cell membranes or silica substrates at physiological pH [
51]. Another feature contributing to the high degree of nonspecific adsorption is its carbohydrate groups, which contain four mannose and three N acetyl glucosamine residues in each monomer. Carbohydrate-binding molecules in the sample matrix can bind specifically to the carbohydrate groups of avidin, limiting its use. Streptavidin, a nonglycosylated avidin, and neutravidin, an engineered commercially available deglycosylated form of avidin, lacks the four mannose and three N acetyl glucosamine residues in each subunit of avidin [
34]. The absence of the carbohydrate moieties make the pI of streptavidin (pI~5–6) and neutravidin (pI~6.3) more acidic, reducing the nonspecific binding [
51] without significantly affecting biotin affinity [
34]. Since the biotin-binding affinity of streptavidin and neutravidin in solution is similar, they are often used interchangeably. It is assumed that streptavidin and neutravidin have interchangeable functionalities due to their similar biotin-binding abilities. However, a study by Sut et al. concluded that the adsorption rates of streptavidin and neutravidin with a biotinylated supported lipid bilayer (SLB) interface differed in a pH-dependent manner, and their attachment to these biotinylated SLB interfaces is very different in terms of attachment kinetics, adlayer properties, and pH sensitivity. This tell us that depending on the application of use of the system, the pI and other relevant interfacial forces in the system must be taken into account when choosing between avidins for a design [
56]. Predicting what product is most appropriate for a given application is complicated by the fact that there are numerous and varied formats of avidins available with no universal method of measurement for biotin binding, making it challenging to accurately compare expectations among products [
57].
Table 2 displays the properties of common avidin derivatives.
Table 2. Properties of avidin derivatives.
| |
Origin |
MW |
pI |
Reference |
| Avidin |
Gallus gallus egg white |
~66–69 kDa |
~10 |
[27] |
| Streptavidin |
Bacterium Streptomyces avidinii |
~56 kDa |
~5–6 |
[27] |
| Neutravidin |
Deglycosylated avidin |
~60 kDa |
~6.3 |
[27] |
| Traptavidin |
S52G, R53D mutant of streptavidin |
~56 kDa |
~5.1 |
[54] |
| Bradavidin II |
Bradyrhizobium japonicum |
~58.4 kDa |
~9.6 |
[27] |
| Tamavidin 2 |
Mushroom Pleurotus cornucopiae |
~ 60.9 kDa |
~7.4 |
[58] |
This entry is adapted from the peer-reviewed paper 10.3390/cimb45110549