Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Cancer is a prime example derived from a loss of homeostasis, primarily caused by genetic alterations both in the genomic and epigenetic landscape, which results in deregulation of the gene networks.
- cancer disease
- lncRNAs
1. Introduction
The Cancer Genome Atlas Consortium project, together with other large-scale sequencing projects aimed at characterizing cancer genomes as well as the possible epigenetic and genetic dysregulations that configure them, have provided a precise molecular characterization of approximately 11,000 primary cancers, discovering a substantial fraction of undescribed somatic abnormalities (e.g., point mutations, genetic rearrangements, and copy number alterations) [1][2][3][4]. Khuruna et al. (2013) reported that 99% of somatic SNVs in different carcinomas occur in non-coding regions (ncRNAs, pseudogenes, and transcription factor binding sites) [5]. Furthermore, a study based on TCGA and lncRNA expression data from TANRIC shows that mutational frequencies in lncRNAs, whose expression is affected by somatic alterations (MutLncs), are low, and that to some extent, their alteration tends to be specific to the type of disease [6].
Interestingly, numerous studies revealed a previously uncovered role for long non-coding RNAs (lncRNAs) as conditional and constitutive oncogenes or tumor suppressor genes through their facility to regulate each and every characteristic of cancer, such as aberrant proliferation, cellular invasion, altered lipid metabolism, metastasis, and immune escape. Furthermore, regulation exerted by lncRNAs can be carried out both at the transcriptional level - epigenetic and genetic - or at the post-transcriptional level [7][8][9][10][11][12]. These findings, and especially the cancer-specific expression of most of them, pointed to lncRNAs as possible biomarkers or therapeutic targets.
2. Genetic and Epigenetic Contributions of Long Non-Coding RNAs Dysregulation in Cancer
The nuclear distribution of lcnRNAs and their interaction with a several nuclear elements, such as transcription factors, chromatin remodeling complexes, or even with a DNA establishing a DNA-RNA complex, have revealed their importance in the regulation of both gene transcription and its epigenetic landscape [13][14][15]. Most types of cancer require numerous changes in nuclear transcription homeostasis in order to escape cellular control systems. These changes translate into upregulation and downregulation of a multitude of genes, oncogenes, and tumor suppressor genes by different mechanisms that result in tumor initiation, progression, and metastasis [16]. In line with this, many studies have pointed out lncRNAs as pivotal modulators of nuclear transcription, altering both the transcriptional and epigenetic cellular context responsible for increasing or repressing tumorogenesis as described below (Figure 1).
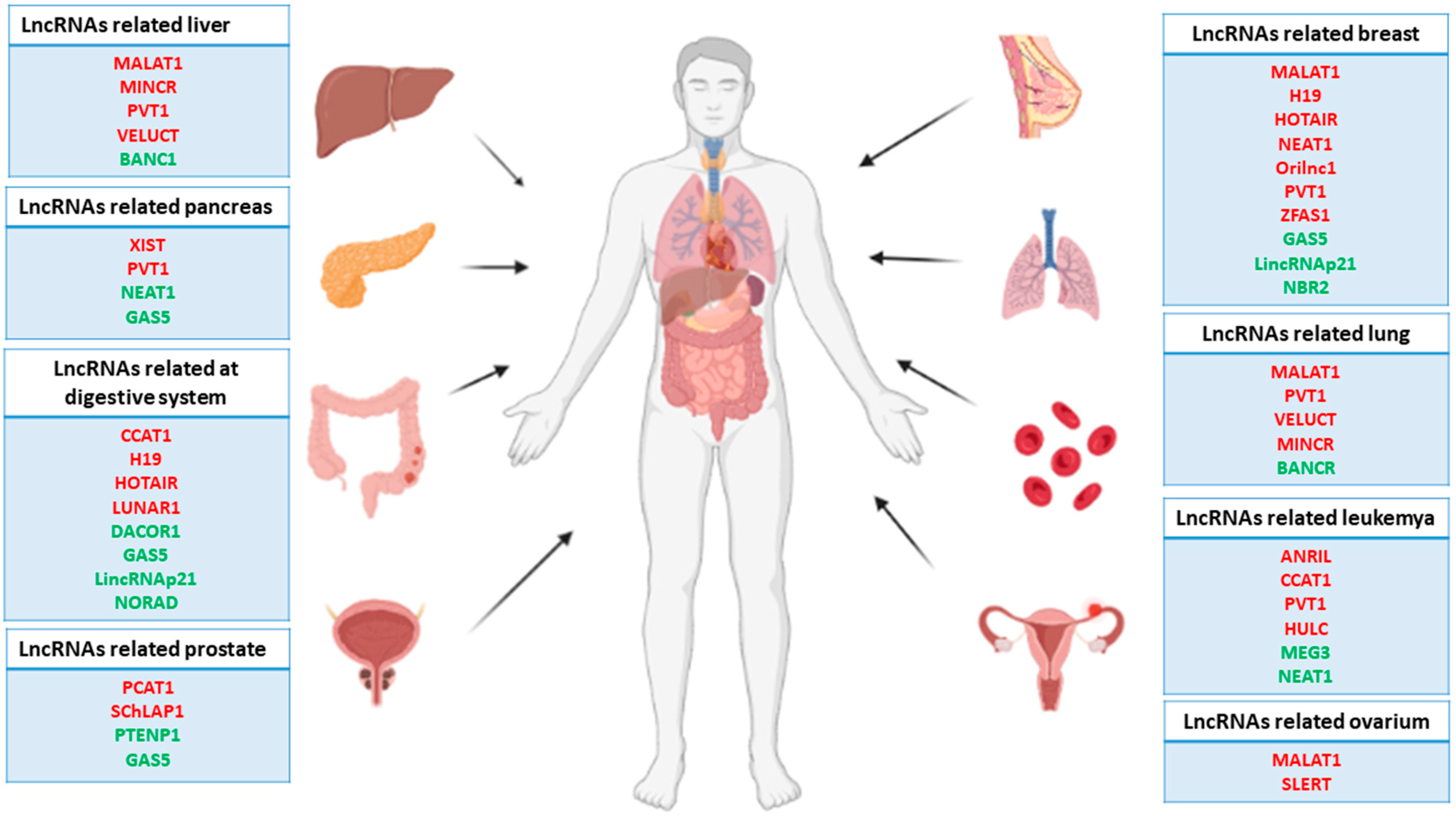
Figure 1. Representative scheme of the main human carcinomas and related lncRNAs. Note that the red lncRNAs act as oncogenes, while the green lncRNAs act as tumor suppressor genes.
Cancer’s underlying epigenetics has been widely studied, given its pivotal role in the initiation, progression, and metastasis of most types of cancer, as well as its potential as a target for different gene therapies [17][18][19]. Epigenetics can be defined as the set of modifications that alters the structure or state of chromatin, differentially regulating its gene expression without changes in the DNA sequence [20]. Epigenetic alterations can be included in two basic groups. The first is modifications in the DNA, basically produced by the addition of methyl groups at the 5′ end of certain cytosines located in the so-called CpG islands, where long stretches of CG dinucleotides harbor the promoter regions of the genome that present high, precise, and intense gene regulation [21]. Methylation of the cytosines prevents binding of the transcription factors to the promoter regions, which results in gene repression. Interestingly, many types of cancer display a specific pattern of hyper-methylated CpG islands in several tumor suppressor gene promotors, inhibiting their expression and leading to an increase in different subpopulation cancer cells and thus enhanced tumor development [22][23][24]. The second group is modifications in the histone tail, essentially caused by methylation, acetylation, or phosphorylation of certain lysines located in the N-terminal region of histones 3′, altering the nucleosome charge, affecting the chromatin state, and thus allowing or preventing the recruitment of transcriptional co-activators to it [25]. These modifications are carried out by chromatin remodeling complexes that add open or close marks to the genome, making it accessible to the transcriptional machinery [26]. Unlike the hyper-methylation of the CpG islands, in many types of cancer, hypo-methylation of the genome is observed, leading to ectopic activation of specific oncogenes, whose expression is inhibited in homeostatic conditions. Ectopic activation of these oncogenes is essential for the cell to acquire an oncogenic phenotype and escape from cellular control systems [27][28]. In the same way, some of these chromatin remodeling complexes show enhanced or reduced activity in malignant cells, which leads to dysregulation in the chromatic structure, promoting the expression of genes that enhance tumor development [29][30][31].
Emerging studies about the role of lncRNAs in carcinogenesis have pointed out as complex regulators in the epigenetics of cancer. As a consequence, many reports of cancer-related lncRNAs have been described as pivotal modulators of the epigenetic landscape by interaction with chromatin at four levels, which are described below: (1) lncRNAs as modulators of histone methylation; (2) lncRNAs as modulators of histone acetylation; (3) lncRNAs as modulators of DNA methylation; (4) lncRNAs as post-transcriptional regulators of the epigenetic apparatus and (5) (Figure 2).
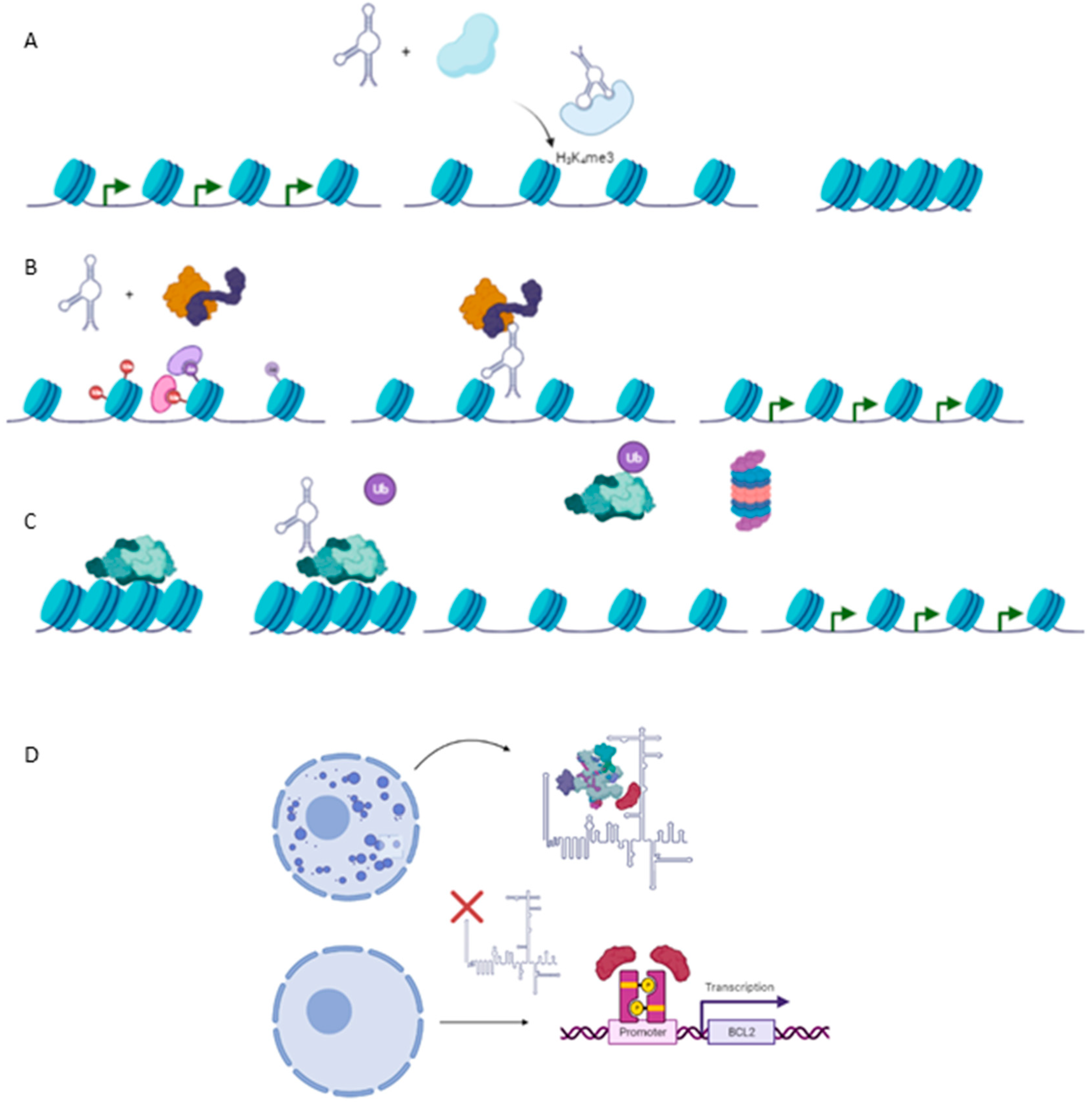
Figure 2. Epigenetic mechanism involved in carcinogenesis. (A) Histone methylation exerted by HOTAIR-PRC2 complex to repress expression of several genes such as Hoxd10, PGR, PCDH, or Jam2 with a protective role against metastasis in breast cancer cell line. (B) TCF21 promoter demethylation exerted by TARID1-GADD45A, which positively modulates expression of the TCF21 gene, a protective factor against head and neck squamous cell carcinoma (HNSCC). (C) Ubiquitination of EZH2, a subunit of PRC2, by ANCR reducing negative marks at Hoxd10 or E-Cadherin genes, exerting a protective role against EMT and metastasis in breast cancer cell line. (D) Required binding SPKQ-Neat1 for the formation of paraspeckles. Downregulation of Neat1 is translated in the high availability of SPKQ and therein the upregulation of apoptosis genes such as BLC2 or BAX.
2.1. Long Non-Coding RNAs as Modulators of Histone Methylation
The interaction between the chromatin remodeling complex PRC2 and HOTAIR was the first reported example of an lncRNA involved in the epigenetic regulation of chromatin [32]. In breast cancer, HOTAIR is necessary to recruit PRC2 to its genomic targets, which is required for the latter to establish the repressor marks H3K27me3 in certain tumor suppressor genes that play pivotal roles in the inhibition of metastasis such as Hoxd10, PGR (progesterone receptor), PCDH (Protocadherin 10) or Jam2 (junctional adhesion molecule). Furthermore, a loss of HOTAIR function is sufficient to prevent cell invasion and metastasis in breast cancer, reflecting the role of this lncRNA as a potent oncogene and pointing it out as a possible therapeutic target [33].
PRC2 subunits can interact with different nuclear proteins or transcription factors, forming protein complexes that establish epigenetic marks on their genomic targets [34]. Tug1, an upregulated lncRNA in different gliomas, acts as a scaffold molecule necessary for PRC2 to interact with YPP1, a neuronal transcription factor. Such a new complex can increase the expression of different genes involved in neuronal differentiation, such as BDNF, NGF, or NFT3, maintaining the pluripotency of malignant cells and thus increasing the aggressiveness of the glioma. Interestingly, the expression of Tug1 is detected in the cytoplasm too, where it acts as a sponge for miR-142, protecting SOX2 and MYC from degradation [35].
Maintenance of the epigenetic marks induced by PRC2 requires the participation of another complex: PRC1 [36]. ANRIL, a long non-coding RNA located in a genetic desert with a high prevalence of SNPs [37][38][39] (to date, many of these SNPs have been related to a higher prevalence of cancer), interacts with both PRC1 through the CBX7 subunit and PRC2 through the SUZ12 subunit to repress the expression of the CDKN2A/B locus and thus maintaining its silence. This locus is downregulated in many types of cancer, such as leukemia, breast cancer, pancreatic cancer, ovarian cancer, and gliomas [40][41][42][43][44]. Repression of the CDNK2A/B locus by ANRIL increases the cell proliferation of malignant cells, promoting progression and metastasis. On the contrary, ANRIL depletion reduces cell proliferation and decides the balance toward cell death, therefore reducing the risk of metastasis, pointing to ANRIL as a potent therapeutic target, especially in leukemia and prostate cancer, where it is upregulated [45][46].
HOTTIP provides another example of lncRNA upregulated in several types of cancer, such as hepatocellular carcinoma, gastric cancer, colorectal cancer, pancreatic cancer, lung cancer, prostate cancer, and osteosarcoma [47][48][49][50][51][52][53]. HOTTIP interacts with WDR5, inducing the chromatic opening through the WDR5/MTT complex. This complex upregulates the expression of the HOXA locus through the H3K4me3 marks. In turn, HOXA locus genes repress the expression of several tumor suppressor genes. Additionally, the detection of HOTTIP in the exosome samples of patients has been recognized as a possible prognosis marker in colorectal cancer [54].
Unlike the lncRNAs described above, MEG3 represents an intergenetic lncRNA associated with chromatin repressive marks that acts as a potent tumor suppressor gene. MEG3 physically interacts with the EZH2 subunit of the PRC2 complex to repress the expression of several genes involved in TFG-β signaling (e.g., SMAD2 or TGFBR1), increasing the aggressiveness of cancer by promoting invasion and metastasis. This repression is mediated by the formation of a triplex DNA-RNA complex in the GA-rich regions of a gene’s silenced promoters [55].
2.2. Long Non-Coding RNAs as Modulators of Histone Acetylation
Few cases of lncRNAs involved in histone acetylation have been described to date. Wan et al. (2013) described a pivotal role of lncRNA JADE in the earliest steps of DNA damage response (DDR) mechanisms by the modulation of acetylation machinery. As suggested in basic and preclinical studies, DDR is one of the primary anti-cancer barriers during tumorigenesis and is under complex and tight regulation, including alteration of the acetylation patterns of numerous gene promoters [56][57][58]. Clinical studies performed in breast cancer patients have shown lncRNA JADE upregulated expression compared with controls. Mechanistically, lncRNA JADE is required for correct JADE1 expression, a protein necessary to determinate the histone H4 substrate specificity of HBO1, which in turn mediates histone H3–H4 acetylation [59]. Several types of cancer display upregulation of HBO1, positively modulating the expression of proliferation-promoting genes linked to a poor cancer prognosis [60][61]. The depletion of lncRNA JADE is translated in the reduced growth of breast cancer in vivo, as well as in HBO1 deficient cells, suggesting a potential effect as inductors of the proliferation of maligned cells [56].
Another case of histone acetylation mediated by lncRNA was reported for lncPRESS1, which plays a pivotal role in the switching of the pluripotent or differentiated state of embryonic stem cells (ESCs) by acting as a decoy molecule from SIRT6. LncPRESS1 competes with SIRT6 for their genomic targets, avoiding that this enzyme can de-acetylate and thus the active gene expression required for promoting cell differentiation [62].
2.3. Long Non-Coding RNAs as Modulators of DNA Methylation
The patterns of aberrant methylation of lncRNA promoters have been described in many types of cancer, pointing out their importance in the epigenetic control of carcinogenesis [63]. However, only a few cases of lncRNAs involved in DNA methylation have been deeply studied, and their functions remain to be fully elucidated. TARID1 is an intergenic lncRNA whose promoter region is located within the third CpG island of the TFC21 promoter, a known tumor suppressor gene. TARID1 binds GADD45A, a DNA repair protein that promotes active demethylation in numerous promoters. The TARID1-GADD45A complex directs it, together with TDG, a protein necessary for GADD45A to interact with its genomic targets, to the TFC21 promoter, where it eliminates methylation and allows for the expression of TFC21, which in turn plays a key role in protection against head and neck squamous cell carcinoma (HNSCC). Interestingly, TARID1 formed an R-loop with the TCF21 promoter, which was recognized by GADD45A as a region marked for demethylation [64][65].
Merry et al. (2015) uncovered a total of 148 lncRNAs that are associated with DNMT1 in colon cancer cells through RIP-seq. Among them, one lncRNA was highlighted, named DNMT1-associated colon cancer repressed lncRNA 1 (DACOR1), which is highly and specifically expressed in normal colon tissue while it is repressed in colon cancer cell lines. Furthermore, overexpression of DACOR1 in colon cancer cells resulted in a gain in DNA methylation at multiple loci without changing the DNMT1 expression level. Interestingly, DNTM1 is an important repressor of tumorigenesis [66]. ChIRP-seq analysis demonstrated that DACOR1 occupies a total of 338 genomic loci, of which 161 peaks are near 150 annotated genes. Remarkably, 31 of these sites overlapped with differentially methylated regions previously found in colon cancer samples with respect to normal tissues. These findings indicate that DACOR1, cooperating with both chromatin and DNMT1, targets the DNMT1 protein complex toward exact genomic loci. Furthermore, DACOR1 was found to repress the expression of cystathionine β-synthase (CBS) and, in turn, increase methionine, which is the substrate to produce S-adenosyl methionine (SAM). SAM is a necessary methyl donor for DNA methylation in mammalian cells. Thus, DACOR1 may also impinge on DNA methylation through orchestrating cellular SAM levels [67][68].
2.4. Long Non-Coding RNAs as Post-Transcriptional Regulators of Related Epigenetic Proteins
The interaction between lncRNAs and the related epigenetic protein landscape is not limited to recruitment, scaffold, or active element functions required for chromatin remodeling. Furthermore, lncRNAs have been reported as pivotal players in modulating the chromatin protein complex stability by promoting protein degradation, exerting protectives roles in most of cases to trigger pro-oncogenic epigenetic marks or inhibit protein degradation, acting as oncogenes. For example, ANCR is capable of directly binding to the EZH2 subunit, promoting its degradation. ANCR-EZH2 binding is required for CDK1 to target EZH2 for ubiquitin–proteasome degradation via Thr-345 and Thr-487 phosphorylation in breast cancer cells. Curiously, in breast cancer, ANCR expression is inactive, leading to hyperactivity of EZH2, which in turn sets up several repressive marks in tumor suppressor genes such as Hoxa10 or E-Cadherin, which are involved in progression and EMT signals [69]. Note that the modulation of ANCR has been probed in other carcinomas [70][71].
EZH2 degradation is not modulated solely by ANCR. MEG3 promotes EZH2 ubiquitination, leading to upregulation of LATS2, a tumor suppressor kinase that inhibits cell proliferation and metastasis through the Hippo signaling pathway in several types of cancer [72]. Gallbladder cancer also displayed a downregulation of MEG3 accompanied by LAST2 downregulation and thus increased cell proliferation and metastasis [73].
Unlike ANCR or MEG3, LUCAT1 is considered an oncogene by protecting the protein degradation of DNMT1 in esophageal squamous cell carcinoma. A depletion in LUCAT1 expression is correlated with reduced levels of DNMT1 expression together with the upregulation of UHRF1, a protein involved in ubiquitination and therefore DNTM1 degradation [74].
2.5. Long Non-Coding RNAs as Nuclear Environment Modulators
The nuclear compartment not only encloses the chromatin in its different phases and the nucleolus but also contains different structures of mostly irregular shapes, known as nuclear bodies [75]. These structures exert pivotal roles in the transcriptional regulation of several pathways related to distinct cellular processes, such as the differentiation, proliferation, or maintenance of homeostasis and, therefore, disease development too [76][77][78][79]. Many reports have highlighted the role of these structures in cancer pathogenesis, involving them in the underlying transcriptional regulation of tumorigenesis [80][81][82]. Along different nuclear bodies, paraspeckles, substructures located on the periphery of the nucleus, were first discovered in 2002 [83] and have emerged as pivotal regulators of nuclear function modulating. First, they distribute nucleus proteins and are available to interact with chromatin or transcriptional machinery [84]. Second, they retain mRNAs, avoiding their transport to the cytoplasm and thereby translation. Interestingly, mRNAs retained by paraspeckles are exported later to the cytoplasm, considerably increasing the number of messenger RNA molecules and their translation. However, the underlying biological processes that determine the export time lapse are poorly understood [85][86]. Third and finally, they sequester proteins related to miRNA biogenesis and processing [87]. The formation of paraspeckles requires the presence of NEAT1, or long non-coding RNA nuclear paraspeckle assembly transcript 1 [88][89]. NEAT1 is transcribed in two isoforms—Neat1.1 and Neat1.2—displaying different RNA processing, which results in the generation of two different transcripts both in their length and in their structural motifs. While the first isoform is dispensable in paraspeckle biogenesis [90], the second isoform is responsible for paraspeckle assembly, constituting a limiting factor for the formation of these nuclear bodies and thus determining the tendency of the nucleus to form them [91]. Zeng et al. (2018) reported the pivotal importance of Neat1.2 and therein paraspeckles as promotors of the aggressiveness of Chronic myeloid leukemia (CML). SPKQ, a bivalent protein that can act as a splicing factor required in paraspeckle formation or as a transcription factor exerting the activation of apoptotic proteins such as BLC2, BBC3, or BAX, is associated with Neat1.2 through the C motif. Neat1.2-SPKQ binding reduces the availability of this protein to act as a transcription factor, thus reducing the expression of apoptosis factors BLC2, BBC3, and BAX and leading the cell to escape apoptosis, thereby enhancing the growth and proliferation of CML. Curiously, Neat1.2 expression is downregulated by c-Myc, a known repressor of CML progression. Neat1.2 repression mediated by c-Myc reduces paraspeckle formation and leads to SPKQ binding to the promoters of the apoptotic genes referred to above, activating their expression and consequently promoting apoptosis and achieving a better prognosis [84].
3. Transcriptional Gene Modulation by Long Non-Coding RNAs
The 3D architecture of the genome is essential for gene transcriptional regulation. RNA polymerases require contact with the promoter regions of the genes to be transcribed [92]. Contact between the distal regions of the genome is facilitated by the transcription factors and active enhancer regions, which generate chromosomal looping. Altering the 3D structure of the chromatin allows RNA polymerases to recognize the distal promoter regions and initiate the synthesis of different transcripts [93]. Active enhancers are indispensable for leading transcription genes that are located far from each other in the genome at the same time [94][95][96]. Under homeostatic conditions, active enhancers control the maintenance of different cell types. However, they are deregulated in many human cancers. Along with them, lncRNAs derived from the transcription of active enhancers are differentially expressed in cancer tissues and have been revealed to act as oncogenes, promoting the transcriptional activation of the oncogenic pathways and even inducing chromosomal rearrangements and genomic instability [97][98][99][100].
The regulatory impact of enhancer-related lncRNAs has been elucidated, especially in human T cell acute lymphoblastic leukemia (T-ALL). Leukemia-induced non-coding activator RNA-1 (LUNAR1) was recognized in an integrated transcriptome profile from T-ALL patients as a specific lncRNA involved in cell growth both in vitro and in vivo in the early stages. LUNAR1 is upregulated by the Notch1/Rbp-jk activator complex, which plays a pivotal role in the initiation of T-ALL carcinogenesis [101][102]. LUNAR1 expression is necessary to promote the IGF1R mRNA levels and to maintain the IGF1 signaling required to stimulate tumor growth. The depletion of LUNAR1 leads to downregulation of IGF1R and a reduction of both the Mediator complex and RNA Pol II binding to both the IGF1R enhancer and promoter [101].
Recently, Tan et al. (2019) described ARIEL, an ARID5B-inducing enhancer associated long non-coding RNA - another example of eRNA involved in T-ALL pathogenesis. The expression of ARIEL is associated with the ARID5B enhancer activity and is required for the recruitment of the Mediator complex toward the ARID5B promoter and thereby increasing the expression of this transcription factor. ARID5B is necessary to activate the TAL1-induced transcriptional program and the MYC oncogene [103]. Curiously, TAL1 positively modulates ARIEL expression, showing a feedback regulatory system. While ARIEL knockdown in cells is translated into growth inhibition, murine model mutants to ARIEL display a block in disease progression, reflecting the importance of this lncRNA in T-ALL pathogenesis and pointing out a possible preventive therapeutic target [104].
This entry is adapted from the peer-reviewed paper 10.3390/ijms23020764
References
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium; Campbell, P.J.; Getz, G. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93.
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Ng, A.W.T.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The repertoire of mutational signatures in human cancer. Nature 2020, 578, 94–101.
- Li, M.; Li, H.; Chen, Q.; Wu, W.; Chen, X.; Ran, L.; Si, G.; Tan, X. A Novel and Robust Long Noncoding RNA Panel to Predict the Prognosis of Pancreatic Cancer. DNA Cell Biol. 2020, 39, 1282–1289.
- Khurana, E.; Fu, Y.; Colonna, V.; Mu, X.J.; Kang, H.M.; Lappalainen, T.; Sboner, A.; Lochovsky, L.; Chen, J.; Harmanci, A.; et al. Integrative Annotation of Variants from 1092 Humans: Application to Cancer Genomics. Science 2013, 342, 1235587.
- Li, J.; Han, L.; Roebuck, P.; Diao, L.; Liu, L.; Yuan, Y.; Weinstein, J.N.; Liang, H. TANRIC: An Interactive Open Platform to Explore the Function of lncRNAs in Cancer. Cancer Res. 2015, 75, 3728–3737.
- Unfried, J.P.; Sangro, P.; Prats-Mari, L.; Sangro, B.; Fortes, P. The Landscape of lncRNAs in Hepatocellular Carcinoma: A Translational Perspective. Cancers 2021, 13, 2651.
- Yan, H.; Chai, H.; Zhao, H. Detecting lncRNA–Cancer Associations by Combining miRNAs, Genes, and Prognosis with Matrix Factorization. Front. Genet. 2021, 12, 639872.
- Vancura, A.; Lanzós, A.; Bosch-Guiteras, N.; Esteban, M.T.; Gutierrez, A.H.; Haefliger, S.; Johnson, R. Cancer LncRNA Census 2 (CLC2): An enhanced resource reveals clinical features of cancer lncRNAs. NAR Cancer 2021, 3, zcab013.
- Ganini, C.; Amelio, I.; Bertolo, R.; Bove, P.; Buonomo, O.C.; Candi, E.; Cipriani, C.; Di Daniele, N.; Juhl, H.; Mauriello, A.; et al. Global mapping of cancers: The Cancer Genome Atlas and beyond. Mol. Oncol. 2021, 15, 2823–2840.
- Liang, Y.; Song, X.; Li, Y.; Chen, B.; Zhao, W.; Wang, L.; Zhang, H.; Liu, Y.; Han, D.; Zhang, N.; et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol. Cancer 2020, 19, 85.
- Xiu, B.; Chi, Y.; Liu, L.; Chi, W.; Zhang, Q.; Chen, J.; Guo, R.; Si, J.; Li, L.; Xue, J.; et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol. Cancer 2019, 18, 1–20.
- Zhang, Y.; Huang, Y.-S.; Wang, D.-L.; Yang, B.; Yan, H.-Y.; Lin, L.-H.; Li, Y.; Chen, J.; Xie, L.-M.; Liao, J.-Y.; et al. LncRNA DSCAM-AS1 interacts with YBX1 to promote cancer progression by forming a positive feedback loop that activates FOXA1 transcription network. Theranostics 2020, 10, 10823–10837.
- Bian, D.; Gao, C.; Bao, K.; Song, G. The long non-coding RNA NKILA inhibits the invasion-metastasis cascade of malignant melanoma via the regulation of NF-ĸB. Am. J. Cancer Res. 2017, 7, 28.
- Vahidi, S.; Norollahi, S.E.; Agah, S.; Samadani, A.A. DNA Methylation Profiling of hTERT Gene Alongside with the Telomere Performance in Gastric Adenocarcinoma. J. Gastrointest. Cancer 2020, 51, 788–799.
- Wang, D.; Ding, L.; Wang, L.; Zhao, Y.; Sun, Z.; Karnes, R.J.; Zhang, J.; Huang, H. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget 2015, 6, 41045–41055.
- Fardi, M.; Solali, S.; Hagh, M.F. Epigenetic mechanisms as a new approach in cancer treatment: An updated review. Genes Dis. 2018, 5, 304–311.
- Richter, A.M.; Woods, M.L.; Küster, M.M.; Walesch, S.K.; Braun, T.; Boettger, T.; Dammann, R.H. RASSF10 is frequently epigenetically inactivated in kidney cancer and its knockout promotes neoplasia in cancer prone mice. Oncogene 2020, 39, 3114–3127.
- He, J.; Zhou, M.; Li, X.; Gu, S.; Cao, Y.; Xing, T.; Chen, W.; Chu, C.; Gu, F.; Zhou, J.; et al. SLC34A2 simultaneously promotes papillary thyroid carcinoma growth and invasion through distinct mechanisms. Oncogene 2020, 39, 2658–2675.
- Holleran, J.L.; Parise, R.A.; Joseph, E.; Eiseman, J.L.; Covey, J.M.; Glaze, E.R.; Lyubimov, A.V.; Chen, Y.-F.; D’Argenio, D.Z.; Egorin, M.J. Plasma Pharmacokinetics, Oral Bioavailability, and Interspecies Scaling of the DNA Methyltransferase Inhibitor, Zebularine. Clin. Cancer Res. 2005, 11, 3862–3868.
- Lacal, I.; Ventura, R. Epigenetic Inheritance: Concepts, Mechanisms and Perspectives. Front. Mol. Neurosci. 2018, 11, 292.
- Scelfo, A.; Fachinetti, D. Keeping the Centromere under Control: A Promising Role for DNA Methylation. Cells 2019, 8, 912.
- Yu, Z.; Feng, J.; Wang, W.; Deng, Z.; Zhang, Y.; Xiao, L.; Wang, Z.; Liu, C.; Liu, Q.; Chen, S.; et al. The EGFR-ZNF263 signaling axis silences SIX3 in glioblastoma epigenetically. Oncogene 2020, 39, 3163–3178.
- Mirmohammadsadegh, A.; Marini, A.; Nambiar, S.; Hassan, M.; Tannapfel, A.; Ruzicka, T.; Hengge, U.R. Epigenetic Silencing of the PTEN Gene in Melanoma. Cancer Res. 2006, 66, 6546–6552.
- Marini, A.; Mirmohammadsadegh, A.; Nambiar, S.; Gustrau, A.; Ruzicka, T.; Hengge, U.R. Epigenetic Inactivation of Tumor Suppressor Genes in Serum of Patients with Cutaneous Melanoma. J. Investig. Dermatol. 2006, 126, 422–431.
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324.
- Magaña-Acosta, M.; Valadez-Graham, V. Chromatin Remodelers in the 3D Nuclear Compartment. Front. Genet. 2020, 11.
- Sastry, N.G.; Wan, X.; Huang, T.; Alvarez, A.A.; Pangeni, R.P.; Song, X.; James, C.D.; Horbinski, C.M.; Brennan, C.W.; Nakano, I. LY6K promotes glioblastoma Tumorigenicity via CAV-1-mediated ERK1/2 signaling enhancement. Mol. Cancer 2020, 19, 1–16.
- Xiao, C.; Wu, G.; Zhou, Z.; Zhang, X.; Wang, Y.; Song, G.; Ding, E.; Sun, X.; Zhong, L.; Li, S.; et al. RBBP6, a RING finger-domain E3 ubiquitin ligase, induces epithelial–mesenchymal transition and promotes metastasis of colorectal cancer. Cell Death Dis. 2019, 10, 1–17.
- Chowdhury, M.; Mihara, K.; Yasunaga, S.; Ohtaki, M.; Takihara, Y.; Kimura, A. Expression of Polycomb-group (PcG) protein BMI-1 predicts prognosis in patients with acute myeloid leukemia. Leukemia 2007, 6, 846–847.
- Frangini, A.; Sjöberg, M.; Roman-Trufero, M.; Dharmalingam, G.; Haberle, V.; Bartke, T.; Lenhard, B.; Malumbres, M.; Vidal, M.; Dillon, N. The Aurora B Kinase and the Polycomb Protein Ring1B Combine to Regulate Active Promoters in Quiescent Lymphocytes. Mol. Cell 2013, 51, 647–661.
- Barbour, H.; Daou, S.; Hendzel, M.; Affar, E.B. Polycomb group-mediated histone H2A monoubiquitination in epigenome regulation and nuclear processes. Nat. Commun. 2020, 11, 1–16.
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323.
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076.
- Van Mierlo, G.; Veenstra, G.J.C.; Vermeulen, M.; Marks, H. The Complexity of PRC2 Subcomplexes. Trends Cell Biol. 2019, 29, 660–671.
- Katsushima, K.; Natsume, A.; Ohka, F.; Shinjo, K.; Hatanaka, A.; Ichimura, N.; Sato, S.; Takahashi, S.; Kimura, H.; Totoki, Y.; et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat. Commun. 2016, 7, 13616.
- Geng, Z.; Gao, Z. Mammalian PRC1 Complexes: Compositional Complexity and Diverse Molecular Mechanisms. Int. J. Mol. Sci. 2020, 21, 8594.
- Huang, X.; Zhang, W.; Shao, Z. Association between long non-coding RNA polymorphisms and cancer risk: A meta-analysis. Biosci. Rep. 2018, 38, BSR20180365.
- Lou, N.; Liu, G.; Pan, Y. Long noncoding RNA ANRIL as a novel biomarker in human cancer. Future Oncol. 2020, 16, 2981–2995.
- Maruei-Milan, R.; Heidari, Z.; Aryan, A.; Asadi-Tarani, M.; Salimi, S. Long non-coding RNA ANRIL polymorphisms in papillary thyroid cancer and its severity. Br. J. Biomed. Sci. 2021, 78, 58–62.
- Zhao, R.; Choi, B.Y.; Lee, M.-H.; Bode, A.M.; Dong, Z. Implications of Genetic and Epigenetic Alterations of CDKN2A (p16 INK4a) in Cancer. EBioMedicine 2016, 8, 30–39.
- Chen, S.; Zhang, J.-Q.; Chen, J.-Z.; Chen, H.-X.; Qiu, F.-N.; Yan, M.-L.; Chen, Y.-L.; Peng, C.-H.; Tian, Y.-F.; Wang, Y.-D. The over expression of long non-coding RNA ANRIL promotes epithelial-mesenchymal transition by activating the ATM-E2F1 signaling pathway in pancreatic cancer: An in vivo and in vitro study. Int. J. Biol. Macromol. 2017, 102, 718–728.
- Wang, C.-H.; Li, Q.-Y.; Nie, L.; Ma, J.; Yao, C.-J.; Chen, F.-P. LncRNA ANRIL promotes cell proliferation, migration and invasion during acute myeloid leukemia pathogenesis via negatively regulating miR-34a. Int. J. Biochem. Cell Biol. 2020, 119, 105666.
- Wang, K.; Hu, Y.-B.; Zhao, Y.; Ye, C. LncRNA ANRIL Regulates Ovarian Cancer Progression and Tumor Stem Cell-Like Characteristics via miR-324-5p/Ran Axis. OncoTargets Ther. 2021, 14, 565–576.
- Sun, L.-Y.; Li, X.-J.; Sun, Y.-M.; Huang, W.; Fang, K.; Han, C.; Chen, Z.-H.; Luo, X.-Q.; Chen, Y.-Q.; Wang, W.-T. LncRNA ANRIL regulates AML development through modulating the glucose metabolism pathway of AdipoR1/AMPK/SIRT. Mol. Cancer 2018, 17, 1–6.
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene 2011, 30, 1956–1962.
- Kong, Y.; Hsieh, C.-H.; Alonso, L.C. ANRIL: A lncRNA at the CDKN2A/B Locus with Roles in Cancer and Metabolic Disease. Front. Endocrinol. 2018, 9, 405.
- Cheng, Y.; Jutooru, I.; Chadalapaka, G.; Corton, J.C.; Safe, S. The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration. Oncotarget 2015, 6, 10840–10852.
- Chang, S.; Liu, J.; Guo, S.; He, S.; Qiu, G.; Lu, J.; Wang, J.; Fan, L.; Zhao, W.; Che, X. HOTTIP and HOXA13 are oncogenes associated with gastric cancer progression. Oncol. Rep. 2016, 35, 3577–3585.
- Tang, Y.; Ji, F. lncRNA HOTTIP facilitates osteosarcoma cell migration, invasion and epithelial-mesenchymal transition by forming a positive feedback loop with c-Myc. Oncol. Lett. 2019, 18, 1649–1656.
- Yang, B.; Gao, G.; Wang, Z.; Sun, D.; Wei, X.; Ma, Y.; Ding, Y. Long non-coding RNA HOTTIP promotes prostate cancer cells proliferation and migration by sponging miR-216a-5p. Biosci. Rep. 2018, 38, BSR20180566.
- Navarro, A.; Moises, J.; Santasusagna, S.; Marrades, R.M.; Viñolas, N.; Castellano, J.J.; Canals, J.; Muñoz, C.; Ramírez, J.; Molins, L.; et al. Clinical significance of long non-coding RNA HOTTIP in early-stage non-small-cell lung cancer. BMC Pulm. Med. 2019, 19, 1–9.
- Liu, K.; Ni, J.; Li, W.; Pan, B.; Yang, Y.; Xia, Q.; Huang, J. The Sp1/FOXC1/HOTTIP/LATS2/YAP/β-catenin cascade promotes malignant and metastatic progression of osteosarcoma. Mol. Oncol. 2020, 14, 2678–2695.
- Liu, T.; Wang, H.; Yu, H.; Bi, M.; Yan, Z.; Hong, S.; Li, S. The Long Non-coding RNA HOTTIP Is Highly Expressed in Colorectal Cancer and Enhances Cell Proliferation and Invasion. Mol. Ther. Nucleic Acids 2020, 19, 612–618.
- Oehme, F.; Krahl, S.; Gyorffy, B.; Muessle, B.; Rao, V.; Greif, H.; Ziegler, N.; Lin, K.; Thepkaysone, M.-L.; Polster, H.; et al. Low level of exosomal long non-coding RNA HOTTIP is a prognostic biomarker in colorectal cancer. RNA Biol. 2019, 16, 1339–1345.
- Mondal, T.; Subhash, S.; Vaid, R.; Enroth, S.; Uday, S.; Reinius, B.; Mitra, S.; Mohammed, A.; James, A.R.; Hoberg, E.; et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA–DNA triplex structures. Nat. Commun. 2015, 6, 1–7, Erratum in Nat. Commun. 2019, 10, 5290.
- Wan, G.; Hu, X.; Liu, Y.; Han, C.; Sood, A.K.; Calin, G.A.; Zhang, X.; Lu, X. A novel non-coding RNA lncRNA-JADE connects DNA damage signalling to histone H4 acetylation. EMBO J. 2013, 32, 2833–2847.
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254.e6.
- Lozano, R.; Castro, E.; Aragón, I.M.; Cendón, Y.; Cattrini, C.; López-Casas, P.P.; Olmos, D. Genetic aberrations in DNA repair pathways: A cornerstone of precision oncology in prostate cancer. Br. J. Cancer 2021, 124, 552–563.
- Han, J.; Lachance, C.; Ricketts, M.D.; McCullough, C.E.; Gerace, M.; Black, B.E.; Cote, J.; Marmorstein, R. The scaffolding protein JADE1 physically links the acetyltransferase subunit HBO1 with its histone H3–H4 substrate. J. Biol. Chem. 2018, 293, 4498–4509.
- Iizuka, M.; Susa, T.; Takahashi, Y.; Tamamori-Adachi, M.; Kajitani, T.; Okinaga, H.; Fukusato, T.; Okazaki, T. Histone acetyltransferase Hbo1 destabilizes estrogen receptor α by ubiquitination and modulates proliferation of breast cancers. Cancer Sci. 2013, 104, 1647–1655.
- Quintela, M.; Sieglaff, D.H.; Gazze, A.S.; Zhang, A.; Gonzalez, D.; Francis, L.; Webb, P.; Conlan, R.S. HBO1 directs histone H4 specific acetylation, potentiating mechano-transduction pathways and membrane elasticity in ovarian cancer cells. Nanomed. Nanotechnol. Biol. Med. 2019, 17, 254–265.
- Jain, A.K.; Xi, Y.; McCarthy, R.; Allton, K.; Akdemir, K.C.; Patel, L.; Aronow, B.; Lin, C.; Li, W.; Yang, L.; et al. LncPRESS1 Is a p53-Regulated LncRNA that Safeguards Pluripotency by Disrupting SIRT6-Mediated De-acetylation of Histone H3K56. Mol. Cell 2016, 64, 967–981.
- Yang, Z.; Xu, F.; Wang, H.; Teschendorff, A.E.; Xie, F.; He, Y. Pan-cancer characterization of long non-coding RNA and DNA methylation mediated transcriptional dysregulation. EBioMedicine 2021, 68, 103399.
- Liu, J.; Jiang, G.; Mao, P.; Zhang, J.; Zhang, L.; Liu, L.; Wang, J.; Owusu, L.; Ren, B.; Tang, Y.; et al. Down-regulation of GADD45A enhances chemosensitivity in melanoma. Sci. Rep. 2018, 8, 1–11.
- Arab, K.; Park, Y.J.; Lindroth, A.; Schäfer, A.; Oakes, C.; Weichenhan, D.; Lukanova, A.; Lundin, E.; Risch, A.; Meister, M.; et al. Long Noncoding RNA TARID Directs Demethylation and Activation of the Tumor Suppressor TCF21 via GADD45A. Mol. Cell 2014, 55, 604–614.
- Wong, K.K. DNMT1 as a therapeutic target in pancreatic cancer: Mechanisms and clinical implications. Cell. Oncol. 2020, 43, 779–792.
- Merry, C.R.; Forrest, M.; Sabers, J.N.; Beard, L.; Gao, X.-H.; Hatzoglou, M.; Jackson, M.W.; Wang, Z.; Markowitz, S.D.; Khalil, A.M. DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Hum. Mol. Genet. 2015, 24, 6240–6253.
- Somasundaram, S.; Forrest, M.E.; Moinova, H.; Cohen, A.; Varadan, V.; LaFramboise, T.; Markowitz, S.; Khalil, A.M. The DNMT1-associated lincRNA DACOR1 reprograms genome-wide DNA methylation in colon cancer. Clin. Epigenet. 2018, 10, 127.
- Li, Z.; Hou, P.; Fan, D.; Dong, M.; Ma, M.; Li, H.; Yao, R.; Li, Y.; Wang, G.; Geng, P.; et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017, 24, 59–71.
- Cai, N.; Li, C.; Wang, F. Silencing of LncRNA-ANCR Promotes the Osteogenesis of Osteoblast Cells in Postmenopausal Osteoporosis via Targeting EZH2 and RUNX2. Yonsei Med. J. 2019, 60, 751–759.
- Wen, Z.; Lian, L.; Ding, H.; Hu, Y.; Xiao, Z.; Xiong, K.; Yang, Q. LncRNA ANCR promotes hepatocellular carcinoma metastasis through upregulating HNRNPA1 expression. RNA Biol. 2020, 17, 381–394.
- Furth, N.; Aylon, Y. The LATS1 and LATS2 tumor suppressors: Beyond the Hippo pathway. Cell Death Differ. 2017, 24, 1488–1501.
- Jin, L.; Cai, Q.; Wang, S.; Wang, S.; Mondal, T.; Wang, J.; Quan, Z. Long noncoding RNA MEG3 regulates LATS2 by promoting the ubiquitination of EZH2 and inhibits proliferation and invasion in gallbladder cancer. Cell Death Dis. 2018, 9, 1–14.
- Yoon, J.-H.; You, B.-H.; Park, C.H.; Kim, Y.J.; Nam, J.-W.; Kil Lee, S. The long noncoding RNA LUCAT1 promotes tumorigenesis by controlling ubiquitination and stability of DNA methyltransferase 1 in esophageal squamous cell carcinoma. Cancer Lett. 2018, 417, 47–57.
- Mao, Y.S.; Zhang, B.; Spector, D.L. Biogenesis and function of nuclear bodies. Trends Genet. 2011, 27, 295–306.
- Elmehdawi, F.; Wheway, G.; Szymanska, K.; Adams, M.; High, A.S.; Johnson, C.A.; Robinson, P.A. Human Homolog of Drosophila Ariadne (HHARI) is a marker of cellular proliferation associated with nuclear bodies. Exp. Cell Res. 2013, 319, 161–172.
- Krejci, J.; Legartová, S.; Bártová, E. Neural Differentiation in HDAC1-Depleted Cells Is Accompanied by Coilin Downregulation and the Accumulation of Cajal Bodies in Nucleoli. Stem. Cells Int. 2017, 2017, 1–13.
- Shastrula, P.K.; Sierra, I.; Deng, Z.; Keeney, F.; Hayden, J.E.; Lieberman, P.M.; Janicki, S.M. PML is recruited to heterochromatin during S phase and represses DAXX-mediated histone H3.3 chromatin assembly. J. Cell Sci. 2019, 132, jcs.220970.
- Ma, F.; Lei, Y.-Y.; Ding, M.-G.; Luo, L.-H.; Xie, Y.-C.; Liu, X.-L. LncRNA NEAT1 Interacted with DNMT1 to Regulate Malignant Phenotype of Cancer Cell and Cytotoxic T Cell Infiltration via Epigenetic Inhibition of p53, cGAS, and STING in Lung Cancer. Front. Genet. 2020, 11, 250.
- Zhu, Y.; Tomlinson, R.L.; Lukowiak, A.A.; Terns, R.M.; Terns, M.P. Telomerase RNA Accumulates in Cajal Bodies in Human Cancer Cells. Mol. Biol. Cell 2004, 15, 81–90.
- Chen, M.; Zhang, J.; Sampieri, K.; Clohessy, J.G.; Mendez, L.; Gonzalez-Billalabeitia, E.; Liu, X.-S.; Lee, Y.-R.; Fung, J.; Katon, J.M.; et al. An aberrant SREBP-dependent lipogenic program promotes metastatic prostate cancer. Nat. Genet. 2018, 50, 206–218.
- Wang, S.; Zhang, Q.; Wang, Q.; Shen, Q.; Chen, X.; Li, Z.; Zhou, Y.; Hou, J.; Xu, B.; Li, N.; et al. NEAT1 paraspeckle promotes human hepatocellular carcinoma progression by strengthening IL-6/STAT3 signaling. OncoImmunology 2018, 7, e1503913.
- Fox, A.H.; Lam, Y.W.; Leung, A.K.; Lyon, C.E.; Andersen, J.; Mann, M.; Lamond, A.I. Paraspeckles: A Novel Nuclear Domain. Curr. Biol. 2002, 12, 13–25.
- Zeng, C.; Liu, S.; Lu, S.; Yu, X.; Lai, J.; Wu, Y.; Chen, S.; Wang, L.; Yu, Z.; Luo, G.; et al. The c-Myc-regulated lncRNA NEAT1 and paraspeckles modulate imatinib-induced apoptosis in CML cells. Mol. Cancer 2018, 17, 1–6.
- Prasanth, K.V.; Prasanth, S.G.; Xuan, Z.; Hearn, S.; Freier, S.M.; Bennett, C.F.; Zhang, M.Q.; Spector, D.L. Regulating Gene Expression through RNA Nuclear Retention. Cell 2005, 123, 249–263.
- Ben-Zvi, M.; Amariglio, N.; Paret, G.; Nevo-Caspi, Y. F11R Expression upon Hypoxia Is Regulated by RNA Editing. PLoS ONE 2013, 8, e77702.
- Jiang, L.; Shao, C.; Wu, Q.-J.; Chen, G.; Zhou, J.; Yang, B.; Li, H.; Gou, L.-T.; Zhang, Y.; Wang, Y.; et al. NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nat. Struct. Mol. Biol. 2017, 24, 816–824.
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007, 8, 1–16.
- Naganuma, T.; Nakagawa, S.; Tanigawa, A.; Sasaki, Y.; Goshima, N.; Hirose, T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012, 31, 4020–4034.
- Li, R.; Harvey, A.R.; Hodgetts, S.I.; Fox, A.H. Functional dissection of NEAT1 using genome editing reveals substantial localization of the NEAT1_1 isoform outside paraspeckles. RNA 2017, 23, 872–881.
- West, J.A.; Mito, M.; Kurosaka, S.; Takumi, T.; Tanegashima, C.; Chujo, T.; Yanaka, K.; Kingston, R.E.; Hirose, T.; Bond, C.; et al. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J. Cell Biol. 2016, 214, 817–830.
- Rowley, J.; Corces, V.G. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 2018, 19, 789–800.
- Savic, D.; Roberts, B.S.; Carleton, J.B.; Partridge, E.C.; White, M.A.; Cohen, B.A.; Cooper, G.M.; Gertz, J.; Myers, R.M. Promoter-distal RNA polymerase II binding discriminates active from inactive CCAAT/enhancer-binding protein beta binding sites. Genome Res. 2015, 25, 1791–1800.
- Spicuglia, S.; Vanhille, L. Chromatin signatures of active enhancers. Nucleus 2012, 3, 126–131.
- Jia, Y.; Chng, W.-J.; Zhou, J. Super-enhancers: Critical roles and therapeutic targets in hematologic malignancies. J. Hematol. Oncol. 2019, 12, 1–17.
- Arnold, P.R.; Wells, A.D.; Li, X.C. Diversity and Emerging Roles of Enhancer RNA in Regulation of Gene Expression and Cell Fate. Front. Cell Dev. Biol. 2020, 7, 377.
- Adeel, M.M.; Jiang, H.; Arega, Y.; Cao, K.; Lin, D.; Cao, C.; Cao, G.; Wu, P.; Li, G. Structural Variations of the 3D Genome Architecture in Cervical Cancer Development. Front. Cell Dev. Biol. 2021, 9, 706375.
- Chen, H.; Liang, H. A High-Resolution Map of Human Enhancer RNA Loci Characterizes Super-enhancer Activities in Cancer. Cancer Cell 2020, 38, 701–715.
- Deng, R.; Huang, J.-H.; Wang, Y.; Zhou, L.-H.; Wang, Z.-F.; Hu, B.-X.; Chen, Y.-H.; Yang, D.; Mai, J.; Li, Z.-L.; et al. Disruption of super-enhancer-driven tumor suppressor gene RCAN1.4 expression promotes the malignancy of breast carcinoma. Mol. Cancer 2020, 19, 1–19.
- Lidschreiber, K.; Jung, L.A.; von der Emde, H.; Dave, K.; Taipale, J.; Cramer, P.; Lidschreiber, M. Transcriptionally active enhancers in human cancer cells. Mol. Syst. Biol. 2021, 17, e9873.
- Trimarchi, T.; Bilal, E.; Ntziachristos, P.; Fabbri, G.; Dalla-Favera, R.; Tsirigos, A.; Aifantis, I. Genome-wide Mapping and Characterization of Notch-Regulated Long Noncoding RNAs in Acute Leukemia. Cell 2014, 158, 593–606.
- Rosati, E.; Baldoni, S.; De Falco, F.; Del Papa, B.; Dorillo, E.; Rompietti, C.; Albi, E.; Falzetti, F.; Di Ianni, M.; Sportoletti, P. NOTCH1 Aberrations in Chronic Lymphocytic Leukemia. Front. Oncol. 2018, 8, 229.
- Wang, P.; Deng, Y.; Yan, X.; Zhu, J.; Yin, Y.; Shu, Y.; Bai, D.; Zhang, S.; Xu, H.; Lu, X. The Role of ARID5B in Acute Lymphoblastic Leukemia and Beyond. Front. Genet. 2020, 11, 598.
- Tan, S.H.; Leong, W.Z.; Ngoc, P.C.T.; Tan, T.K.; Bertulfo, F.C.; Lim, M.C.; An, O.; Li, Z.; Yeoh, A.E.J.; Fullwood, M.J.; et al. The enhancer RNA ARIEL activates the oncogenic transcriptional program in T-cell acute lymphoblastic leukemia. Blood 2019, 134, 239–251.
This entry is offline, you can click here to edit this entry!
