The target of rapamycin (TOR) is an evolutionarily-conserved serine/threonine kinase that senses and integrates signals from the environment to coordinate developmental and metabolic processes. In plants, TOR has been shown to be a central regulator of growth and a negative regulator of catabolic processes such as autophagy.
- autophagy
- plant growth
- target of rapamycin (TOR)
- TORC1
- TOR signaling
1. Introduction
All living organisms require continuous monitoring and integration of signals from their environments for survival, growth, and homeostasis. The target of rapamycin (TOR) kinase complex, an evolutionarily-conserved serine/threonine protein kinase, senses and integrates environmental cues, such as nutrients and stress signals [1][2], and internal signals, like energy and hormones [3][4], to regulate growth and metabolism in both photosynthetic and non-photosynthetic organisms.
Our understanding of TOR signaling, and cellular function began several decades ago when two TOR genes were first isolated from the budding yeast Saccharomyces cerevisiae [5][6]. Subsequently, TOR signaling, function, and structure have been elucidated in other non-photosynthetic organisms such as fungi, Drosophila, and mammals [1][7][8][9][10], and photosynthetic organisms including Arabidopsis thaliana and algae [11][12]]. In fungi and metazoans, TOR forms two structurally and physiologically distinct complexes, termed TORC1 and TORC2 (TOR complex 1 and 2), which regulate cell growth and metabolism in space and time [6][13][14]. To date, there is no evidence for the existence of TORC2-related proteins in plants or microalgae [2][61], suggesting evolutionary conservation of only TORC1 and therefore we discuss only TORC1 and related proteins here.
Cellular anabolic and catabolic processes must both be regulated to maintain homeostasis during growth. Given nutrient-replete or abundant energy conditions, TOR kinase is active and promotes anabolic processes leading to growth [3][15]. Conversely, TOR activity is inhibited in response to stress conditions or upon treatment with inhibitors such as rapamycin, leading to the activation of catabolic processes such as autophagy [16][17][18][19].
Autophagy is a stress-responsive process found throughout eukaryotes by which cell components, including macromolecules and malfunctioning or damaged organelles, are degraded and constituents recycled. Three functionally similar but mechanistically distinct forms of autophagy, including macroautophagy [20][21][22][23][24] and microautophagy [25][26][27][28] in animals, plants and yeast, and chaperone-mediated autophagy in animals [29][30], have been described. In this review, we focus on the most studied and well-characterized form, macroautophagy (hereafter referred to as autophagy) [31][32][33][34][25][26][27][28][29][30][31][32][33][34][35][36][37][38][39][40][41][42][43] and the role of TORC1 in autophagy regulation.
Under normal nutrient-rich conditions, autophagy is maintained at low basal levels (basal autophagy) and its disruption may result in impaired homeostasis and abnormal conditions such as cancer and neurodegenerative disorders [44][45][46]. The low basal autophagy level is maintained by TOR-mediated phosphorylation of ATG13, preventing autophagy induction by the ATG1-ATG13 kinase complex [47][48][49][50]. However, stress conditions such as nutrient starvation and cellular energy deficit [19][51][52][53][54][55][56] trigger responses that reduce TOR activity, which in turn leads to autophagy induction. Although we focus on general non-selective autophagy, it is noteworthy that autophagy can be selective, and targeting of specific cell organelles and macromolecules to the lysosome/vacuole for degradation has been described in detail elsewhere [57][58][59][60].
2. TOR Complex Components
2.1. TORC1 Components
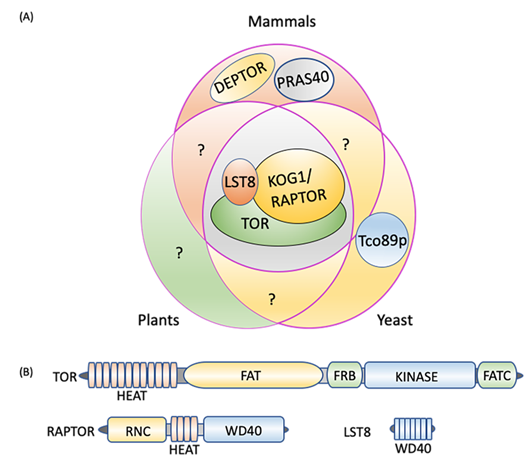
In yeast, plants, and animals, TORC1 constitutes a catalytic subunit, TOR itself, a regulatory subunit, LST8 (lethal with sec thirteen 8), and RAPTOR (regulatory-associated protein of TOR/kontroller of growth 1 (KOG1) in yeast) [62][63][64] (Figure 1A). Yeast-specific core components include Tco89p (89 kDa protein of TOR complex one), which is absent in mammals and plants [6][65], while the additional components of the mammalian TORC1 are DEPTOR (DEP-domain-containing mTOR-interacting protein), [66], and PRAS40 (40 kDa pro-Rich AKT substrate) [67] (Figure 1A). DEPTOR and PRAS40 have inhibitory activity toward mammalian TORC1 and regulate substrate recruitment and apoptosis in cancer cells [66][68][69][70]. Whereas these subunits appear to be absent in yeast and plants, it is possible that they are present but diverged in sequence and if this is the case, could they add yet another regulatory switch to TOR-autophagy signaling? For instance, these TORC1 components may activate autophagy by inhibiting TORC1 even in nutrient sufficient conditions in which autophagy is normally downregulated. Future research may uncover these or related components in yeast and plants and help decipher other autophagy regulatory mechanisms.
Figure 1. TORC1 composition. (A) Three core components, TOR, LST8, and RAPTOR/KOG1, are conserved in plants, mammals, and yeast. DEPTOR and PRAS40 are unique to mammals and Tco89p to yeast. (B) TORC1 protein domains. TOR consists of five distinct domains, the N-terminal HEAT repeats, FAT, FRB, kinase, and the C-terminal FATC. RAPTOR is composed of an N-terminal RNC domain, HEAT repeats in the middle section and a WD40 domain at the C-terminus, while LST8 is made up of seven WD40 domains. DEPTOR: DEP domain containing mTOR interacting protein; FAT: focal adhesion target/FRAP-ATM-TTRAP domain; FATC: FAT-carboxy terminal domain; FRB: FKBP12-rapamycin binding domain; HEAT: Huntingtin elongation factor 3-regulatory subunit A of PP2A-TOR1 repeats; KOG1: kontroller of growth 1; LST8: lethal with Sec13 protein 8; PRAS40: proline-rich Akt substrate of 40 kDa; RAPTOR: regulatory protein associated with TOR; RNC: RAPTOR N-terminus conserved domain; Tco89p: 89 kDa protein of TOR complex one; TOR: target of rapamycin; WD40: tryptophan-aspartic acid repeats of 40 amino acids.
2.2. TORC1 Protein Domain Structures
The TOR protein kinase catalytic subunit consists of conserved N-terminal HEAT (Huntington-EF3-PP2A-TOR1) repeats (Figure 1B), which interact with RAPTOR and are required for substrate recruitment and membrane association. In mammals, the localization of TORC1 to the lysosome membrane is necessary for its negative regulation of autophagy when nutrients, particularly amino acids, are abundant [63][71][72] (see Section 3 for details). By contrast, the yeast TORC1 appears to constitutively localize to the vacuole membrane [73] whereas subcellular localization of plant TORC1 under different conditions needs further research. In the central region of TOR lies a focal adhesion target (FAT) domain potentially involved in interactions with partner proteins [64][74] and which contributes to kinase activation, a necessary step for TORC1 signaling. The catalytic Ser/Thr kinase domain is sandwiched between the FKBP12/rapamycin binding (FRB) domain, and the FATC (FAT C-terminus) domain at the extreme C-terminus. The FRB domain is the binding site for the FKBP12-rapamycin complex [5][6][61][75][76], while the FATC domain serves a scaffolding and kinase activation role [77].
RAPTOR is made up of a RAPTOR N-terminal conserved (RNC) domain, a HEAT domain, and a WD40 (tryptophan-aspartic acid repeats of 40 amino acids) domain at the C-terminus [78][79] (Figure 1B). RAPTOR binds to the HEAT repeat of TOR, selectively presents substrates to the kinase domain, and facilitates substrate phosphorylation [2][80][81][82][83]. RAPTOR also has a role in assembly and stabilization of the complex in mammals [78][79], and the conserved nature of the RNC and HEAT repeats in RAPTOR orthologs suggests a conserved role in other organisms. In Arabidopsis, knock-out of RAPTOR activates autophagy under non-stressed conditions, suggesting that RAPTOR plays a role in TOR-mediated autophagy repression [19]. LST8 is mainly composed of WD40 repeats, like the β-propeller fold of WD-repeat proteins [61][84] (Figure 1B). By interacting with the C-terminal domain of the catalytic subunit, LST8 contributes to TOR complex stability and catalytic activity regulation in yeast, plants, and animals [85][86][87][88].
The existence of TORC1 as a multiprotein complex suggests different possibilities/avenues of signal integration and control. Given that cells experience constant fluctuations in the internal and external environment, it is plausible that a key protein such as TORC1 must adapt quickly and rapidly to these changes to maintain cellular function, and this can be achieved through regulation of different components. Moreover, could these subunits be involved in sensing of upstream signals, or could they mediate TOR interaction with other proteins to regulate autophagy? How does RAPTOR select the substrates to be presented to TOR? Could it have substrate-specific interacting motifs? Or is the structure modified for substrate binding? Future research may aid in answering these or similar questions to fully understand the cooperative processes by which TOR and associated proteins integrate different signals to regulate autophagy. For instance, insight could be generated from structural studies of TORC1 using cryo- electron microscopy (Cryo-EM), for example, although a detailed structure has been generated for mammalian TORC1, more information is needed for plant TORC1 to provide a mechanistic understanding of TOR-autophagy signaling.
This entry is adapted from the peer-reviewed paper 10.3390/ijms21218259
References
- González, A.; Hall, M.N. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017, 36, 397–408.
- Dobrenel, T.; Caldana, C.; Hanson, J.; Robaglia, C.; Vincentz, M.; Veit, B.; Meyer, C. TOR signaling and nutrient sensing. Rev. Plant Biol. 2016, 67, 261–285.
- Wullschleger, S.; Loewith, R.; Hall, M.N. TOR signaling in growth and metabolism. Cell 2006, 124, 471–484.
- Fu, L.; Wang, P.; Xiong, Y. Target of rapamycin signaling in plant stress responses. Plant Physiol. 2020, 182, 1613–1623.
- Heitman, J.; Movva, N.R.; Hall, M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991, 253, 905–909.
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Cell 2002, 10, 457–468.
- Brown, E.J.; Albers, M.W.; Shin, T.B.; Ichikawa, K.; Keith, C.T.; Lane, W.S.; Schreiber, S.L. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 1994, 369, 756–758.
- Cruz, M.C.; Cavallo, L.M.; Görlach, J.M.; Cox, G.; Perfect, J.R.; Cardenas, M.E.; Heitman, J. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in cryptococcus neoformans. Cell Biol. 1999, 19, 4101–4112.
- Oldham, S.; Montagne, J.; Radimerski, T.; Thomas, G.; Hafen, E. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 2000, 14, 2689–2694.
- Cruz, M.C.; Goldstein, A.L.; Blankenship, J.; Del Poeta, M.; Perfect, J.R.; McCusker, J.H.; Bennani, Y.L.; Cardenas, M.E.; Heitman, J. Rapamycin and less immunosuppressive analogs are toxic to candida albicans and cryptococcus neoformans via FKBP12-dependent inhibition of TOR. Agents Chemother. 2001, 45, 3162–3170.
- Menand, B.; Desnos, T.; Nussaume, L.; Berger, F.; Bouchez, D.; Meyer, C.; Robaglia, C. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Natl. Acad. Sci. USA 2002, 99, 6422–6427.
- Crespo, J.L.; Díaz-Troya, S.; Florencio, F.J. Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii. Plant Physiol. 2005, 139, 1736–1749.
- Montero, J.C.; Chen, X.; Ocaña, A.; Pandiella, A. Predominance of mTORC1 over mTORC2 in the regulation of proliferation of ovarian cancer cells: Therapeutic implications. Cancer Ther. 2012, 11, 1342–1352.
- Eltschinger, S.; Loewith, R. TOR complexes and the maintenance of cellular homeostasis. Trends Cell Biol. 2016, 26, 148–159.
- Barbet, N.C.; Schneider, U.; Helliwell, S.B.; Stansfield, I.; Tuite, M.F.; Hall, M.N. TOR controls translation initiation and early G1 progression in yeast. Biol. Cell 1996, 7, 25–42.
- Wang, Y.; Zhang, H. Regulation of autophagy by mTOR signaling pathway. Exp. Med. Biol. 2019, 1206, 67–83.
- Ren, M.; Venglat, P.; Qiu, S.; Feng, L.; Cao, Y.; Wang, E.; Xiang, D.; Wang, J.; Alexander, D.; Chalivendra, S.; et al. Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell 2012, 24, 4850–4874.
- Díaz-Troya, S.; Pérez-Pérez, M.E.; Florencio, F.J.; Crespo, J.L. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 2008, 4, 851–865.
- Pu, Y.; Luo, X.; Bassham, D.C. TOR-dependent and -independent pathways regulate autophagy in Arabidopsis thaliana. Plant Sci. 2017, 8, 1204.
- Juhasz, G.; Neufeld, T.P. Autophagy: A forty-year search for a missing membrane source. PLoS Biol. 2006, 4, e36.
- Yang, Z.; Klionsky, D.J. Mammalian autophagy: Core molecular machinery and signaling regulation. Opin. Cell Biol. 2010, 22, 124–131.
- Lv, X.; Pu, X.; Qin, G.; Zhu, T.; Lin, H. The roles of autophagy in development and stress responses in Arabidopsis thaliana. Apoptosis 2014, 19, 905–921.
- Wen, X.; Klionsky, D.J. An overview of macroautophagy in yeast. Mol. Biol. 2016, 428, 1681–1699.
- McPhee, C.K.; Baehrecke, E.H. Autophagy in Drosophila melanogaster. Biophys. Acta 2009, 1793, 1452–1460.
- Bassham, D.C.; Laporte, M.; Marty, F.; Moriyasu, Y.; Ohsumi, Y.; Olsen, L.J.; Yoshimoto, K. Autophagy in development and stress responses of plants. Autophagy 2006, 2, 2–11.
- Sieńko, K.; Poormassalehgoo, A.; Yamada, K.; Goto-Yamada, S. Microautophagy in plants: Consideration of its molecular mechanism. Cells 2020, 9, 887.
- Mijaljica, D.; Prescott, M.; Devenish, R.J. Microautophagy in mammalian cells: Revisiting a 40-year-old conundrum. Autophagy 2011, 7, 673–682.
- Oku, M.; Sakai, Y. Three distinct types of microautophagy based on membrane dynamics and molecular machineries. Bioessays 2018, 40, e1800008.
- Hao, Y.; Kacal, M.; Ouchida, A.T.; Zhang, B.; Norberg, E.; Vakifahmetoglu-Norberg, H. Targetome analysis of chaperone-mediated autophagy in cancer cells. Autophagy 2019, 15, 1558–1571.
- Dice, J.F. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem. Sci. 1990, 15, 305–309.
- Qi, H.; Xia, F.N.; Xiao, S. Autophagy in plants: Physiological roles and post-translational regulation. Integr Plant Biol. 2020, in press.
- Su, T.; Li, X.; Yang, M.; Shao, Q.; Zhao, Y.; Ma, C.; Wang, P. Autophagy: An intracellular degradation pathway regulating plant survival and stress response. Plant Sci. 2020, 11, 164.
- Ravanan, P.; Srikumar, I.F.; Talwar, P. Autophagy: The spotlight for cellular stress responses. Life Sci. 2017, 188, 53–67.
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836.
- Lamb, C.A.; Yoshimori, T.; Tooze, S.A. The autophagosome: Origins unknown, biogenesis complex. Rev. Mol. Cell Biol. 2013, 14, 759–774.
- Parzych, K.R.; Klionsky, D.J. An overview of autophagy: Morphology, mechanism, and regulation. Redox Signal. 2014, 20, 460–473.
- Yang, Z.; Klionsky, D.J. An overview of the molecular mechanism of autophagy. Top. MicroBiol. Immunol. 2009, 335, 1–32.
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873.
- Liu, Y.; Bassham, D.C. Autophagy: Pathways for self-eating in plant cells. Rev. Plant Biol. 2012, 63, 215–237.
- Ylä-Anttila, P.; Vihinen, H.; Jokitalo, E.; Eskelinen, E.L. 3d tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 2009, 5, 1180–1185.
- Le Bars, R.; Marion, J.; Le Borgne, R.; Satiat-Jeunemaitre, B.; Bianchi, M.W. ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Commun. 2014, 5, 4121.
- Zhuang, X.; Chung, K.P.; Cui, Y.; Lin, W.; Gao, C.; Kang, B.H.; Jiang, L. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Natl. Acad. Sci. USA 2017, 114, E426–E435.
- He, C.; Klionsky, D.J. Regulation mechanisms and signaling pathways of autophagy. Rev. Genet. 2009, 43, 67–93.
- Musiwaro, P.; Smith, M.; Manifava, M.; Walker, S.A.; Ktistakis, N.T. Characteristics and requirements of basal autophagy in hek 293 cells. Autophagy 2013, 9, 1407–1417.
- Cai, Y.; Arikkath, J.; Yang, L.; Guo, M.L.; Periyasamy, P.; Buch, S. Interplay of endoplasmic reticulum stress and autophagy in neurodegenerative disorders. Autophagy 2016, 12, 225–244.
- Davidson, S.M.; Vander Heiden, M.G. Critical functions of the lysosome in cancer biology. Rev. Pharmacol. Toxicol. 2017, 57, 481–507.
- Stephan, J.S.; Yeh, Y.Y.; Ramachandran, V.; Deminoff, S.J.; Herman, P.K. The TOR and PKA signaling pathways independently target the ATG1/ATG13 protein kinase complex to control autophagy. Natl. Acad. Sci. USA 2009, 106, 17049–17054.
- Puente, C.; Hendrickson, R.C.; Jiang, X. Nutrient-regulated phosphorylation of ATG13 inhibits starvation-induced autophagy. Biol. Chem. 2016, 291, 6026–6035.
- Wallot-Hieke, N.; Verma, N.; Schlütermann, D.; Berleth, N.; Deitersen, J.; Böhler, P.; Stuhldreier, F.; Wu, W.; Seggewiß, S.; Peter, C.; et al. Systematic analysis of ATG13 domain requirements for autophagy induction. Autophagy 2018, 14, 743–763.
- Kapahi, P.; Chen, D.; Rogers, A.N.; Katewa, S.D.; Li, P.W.; Thomas, E.L.; Kockel, L. With TOR, less is more: A key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010, 11, 453–465.
- Wang, P.; Zhao, Y.; Li, Z.; Hsu, C.C.; Liu, X.; Fu, L.; Hou, Y.J.; Du, Y.; Xie, S.; Zhang, C.; et al. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Cell 2018, 69, 100–112.e106.
- Lee, J.W.; Park, S.; Takahashi, Y.; Wang, H.G. The association of AMPK with ULK1 regulates autophagy. PLoS ONE 2010, 5, e15394.
- Zhang, Z.; Zhu, J.Y.; Roh, J.; Marchive, C.; Kim, S.K.; Meyer, C.; Sun, Y.; Wang, W.; Wang, Z.Y. TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Biol. 2016, 26, 1854–1860.
- Das, R.; Melo, J.A.; Thondamal, M.; Morton, E.A.; Cornwell, A.B.; Crick, B.; Kim, J.H.; Swartz, E.W.; Lamitina, T.; Douglas, P.M.; et al. The homeodomain-interacting protein kinase HPK-1 preserves protein homeostasis and longevity through master regulatory control of the HSF-1 chaperone network and TORC1-restricted autophagy in Caenorhabditis elegans. PLoS Genet. 2017, 13, e1007038.
- Dunlop, E.A.; Tee, A.R. mTOR and autophagy: A dynamic relationship governed by nutrients and energy. Cell Dev. Biol. 2014, 36, 121–129.
- González, A.; Hall, M.N.; Lin, S.C.; Hardie, D.G. AMPK and TOR: The yin and yang of cellular nutrient sensing and growth control. Cell Metab. 2020, 31, 472–492.
- Evans, T.D.; Sergin, I.; Zhang, X.; Razani, B. Target acquired: Selective autophagy in cardiometabolic disease. Signal. 2017, 10, eaag2298.
- Kazibwe, Z.; Liu, A.Y.; MacIntosh, G.C.; Bassham, D.C. The ins and outs of autophagic ribosome turnover. Cells 2019, 8, 1603.
- Farré, J.C.; Subramani, S. Mechanistic insights into selective autophagy pathways: Lessons from yeast. Rev. Mol. Cell Biol. 2016, 17, 537–552.
- Stephani, M.; Dagdas, Y. Plant selective autophagy-still an uncharted territory with a lot of hidden gems. Mol. Biol. 2020, 432, 63–79.
- Díaz-Troya, S.; Florencio, F.J.; Crespo, J.L. Target of rapamycin and LST8 proteins associate with membranes from the endoplasmic reticulum in the unicellular green alga chlamydomonas reinhardtii. Eukaryot Cell 2008, 7, 212–222.
- Takahara, T.; Maeda, T. Evolutionarily conserved regulation of TOR signalling. Biochem. 2013, 154, 1–10.
- Kim, D.H.; Sarbassov, D.D.; Ali, S.M.; King, J.E.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. mTOR interacts with RAPTOR to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002, 110, 163–175.
- Maegawa, K.; Takii, R.; Ushimaru, T.; Kozaki, A. Evolutionary conservation of TORC1 components, TOR, RAPTOR, and LST8, between rice and yeast. Genet. Genom. 2015, 290, 2019–2030.
- Reinke, A.; Anderson, S.; McCaffery, J.M.; Yates, J.; Aronova, S.; Chu, S.; Fairclough, S.; Iverson, C.; Wedaman, K.P.; Powers, T. TOR complex 1 includes a novel component, TCO89p (YPL180w), and cooperates with SSD1p to maintain cellular integrity in Saccharomyces cerevisiae. Biol. Chem. 2004, 279, 14752–14762.
- Peterson, T.R.; Laplante, M.; Thoreen, C.C.; Sancak, Y.; Kang, S.A.; Kuehl, W.M.; Gray, N.S.; Sabatini, D.M. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 2009, 137, 873–886.
- Sancak, Y.; Thoreen, C.C.; Peterson, T.R.; Lindquist, R.A.; Kang, S.A.; Spooner, E.; Carr, S.A.; Sabatini, D.M. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Cell 2007, 25, 903–915.
- Wang, L.; Harris, T.E.; Roth, R.A.; Lawrence, J.C. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. Biol. Chem. 2007, 282, 20036–20044.
- Thedieck, K.; Polak, P.; Kim, M.L.; Molle, K.D.; Cohen, A.; Jenö, P.; Arrieumerlou, C.; Hall, M.N. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS ONE 2007, 2, e1217.
- Vander Haar, E.; Lee, S.I.; Bandhakavi, S.; Griffin, T.J.; Kim, D.H. Insulin signalling to mTOR mediated by the AKT/PBK substrate pRAS40. Cell Biol. 2007, 9, 316–323.
- Kim, E.; Goraksha-Hicks, P.; Li, L.; Neufeld, T.P.; Guan, K.L. Regulation of TORC1 by RAG GTPases in nutrient response. Cell Biol. 2008, 10, 935–945.
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The RAG GTPases bind RAPTOR and mediate amino acid signaling to mTORC. Science 2008, 320, 1496–1501.
- Binda, M.; Péli-Gulli, M.P.; Bonfils, G.; Panchaud, N.; Urban, J.; Sturgill, T.W.; Loewith, R.; De Virgilio, C. The VAM6 GEF controls TORC1 by activating the EGO complex. Cell 2009, 35, 563–573.
- Watanabe, R.; Wei, L.; Huang, J. mTOR signaling, function, novel inhibitors, and therapeutic targets. Nucl. Med. 2011, 52, 497–500.
- Pérez-Pérez, M.E.; Florencio, F.J.; Crespo, J.L. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 2010, 152, 1874–1888.
- Choi, J.; Chen, J.; Schreiber, S.L.; Clardy, J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 1996, 273, 239–242.
- Tatebe, H.; Shiozaki, K. Evolutionary conservation of the components in the TOR signaling pathways. Biomolecules 2017, 7, 77.
- Aylett, C.H.; Sauer, E.; Imseng, S.; Boehringer, D.; Hall, M.N.; Ban, N.; Maier, T. Architecture of human mTOR complex 1. Science 2016, 351, 48–52.
- Yang, H.; Wang, J.; Liu, M.; Chen, X.; Huang, M.; Tan, D.; Dong, M.Q.; Wong, C.C.; Xu, Y.; Wang, H.W. 4.4 å resolution cryo-em structure of human mTOR complex 1. Protein Cell 2016, 7, 878–887.
- Schalm, S.S.; Fingar, D.C.; Sabatini, D.M.; Blenis, J. TOS motif-mediated RAPTOR binding regulates 4E-BP1 multisite phosphorylation and function. Biol. 2003, 13, 797–806.
- Dunlop, E.A.; Dodd, K.M.; Seymour, L.A.; Tee, A.R. Mammalian target of rapamycin complex 1-mediated phosphorylation of eukaryotic initiation factor 4E-binding protein 1 requires multiple protein-protein interactions for substrate recognition. Cell Signal. 2009, 21, 1073–1084.
- Schalm, S.S.; Blenis, J. Identification of a conserved motif required for mTOR signaling. Biol. 2002, 12, 632–639.
- Mahfouz, M.M.; Kim, S.; Delauney, A.J.; Verma, D.P. Arabidopsis target of rapamycin interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 2006, 18, 477–490.
- Smith, T.F.; Gaitatzes, C.; Saxena, K.; Neer, E.J. The WD repeat: A common architecture for diverse functions. Trends BioChem. Sci. 1999, 24, 181–185.
- Yang, H.; Rudge, D.G.; Koos, J.D.; Vaidialingam, B.; Yang, H.J.; Pavletich, N.P. mTOR kinase structure, mechanism and regulation. Nature 2013, 497, 217–223.
- Kim, D.H.; Sarbassov, D.D.; Ali, S.M.; Latek, R.R.; Guntur, K.V.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. GBETAL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between RAPTOR and mTOR. Cell 2003, 11, 895–904.
- Schepetilnikov, M.; Ryabova, L.A. Recent discoveries on the role of TOR (target of rapamycin) signaling in translation in plants. Plant Physiol. 2018, 176, 1095–1105.
- Wullschleger, S.; Loewith, R.; Oppliger, W.; Hall, M.N. Molecular organization of target of rapamycin complex 2. Biol. Chem. 2005, 280, 30697–30704.