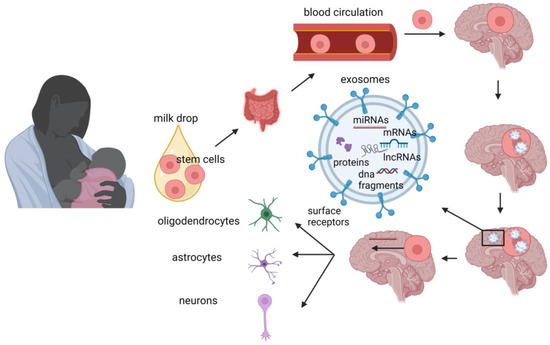
The advantages of human milk feeding, especially in preterm babies, are well recognized. Infants’ feeding with breast milk lowers the likelihood of developing a diverse range of non-communicable diseases later in life and it is also associated with improved neurodevelopmental outcomes. Although the precise mechanisms through which human milk feeding is linked with infants’ neurodevelopment are still unknown, potential epigenetic effects of breast milk through its bioactive components, including non-coding RNAs, stem cells and microbiome, could at least partly explain this association. Micro- and long-non-coding RNAs, enclosed in milk exosomes, as well as breast milk stem cells, survive digestion, reach the circulation and can cross the blood–brain barrier. Certain non-coding RNAs potentially regulate genes implicated in brain development and function, whereas nestin-positive stem cells can possibly differentiate into neural cells or/and act as epigenetic regulators in the brain.
- epigenetics
- human milk
- neurodevelopment
- miRNAs
- long non-coding RNAs
- stem cells
- microbiome
1. Introduction
2. MiRNAs
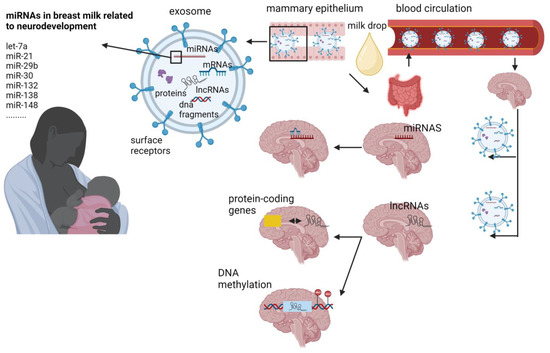
3. Long Non-Coding RNAs
4. Stem Cells
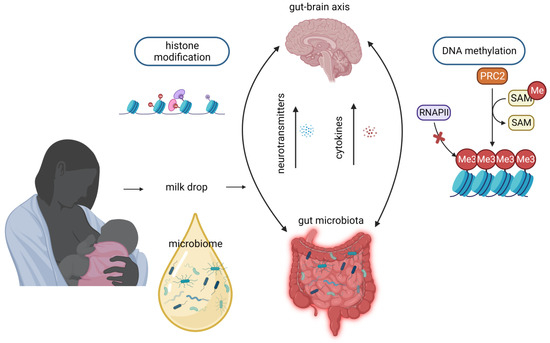
5. Microbiome
This entry is adapted from the peer-reviewed paper 10.3390/nu15163614
References
- GBD 2015 Obesity Collaborators; Afshin, A.; Forouzanfar, M.H.; Reitsma, M.B.; Sur, P.; Estep, K.; Lee, A.; Marczak, L.; Mokdad, A.H.; Moradi-Lakeh, M.; et al. Health Effects of Overweight and Obesity in 195 Countries over 25 Years. N. Engl. J. Med. 2017, 377, 13–27.
- Wilson, J. The Barker hypothesis. An analysis. Aust. N. Z. J. Obstet. Gynaecol. 1999, 39, 1–7.
- Calkins, K.; Devaskar, S.U. Fetal origins of adult disease. Curr. Probl. Pediatr. Adolesc. Health Care 2011, 41, 158–176.
- Barker, D.J.; Osmond, C.; Kajantie, E.; Eriksson, J.G. Growth and chronic disease: Findings in the Helsinki Birth Cohort. Ann. Hum. Biol. 2009, 36, 445–458.
- Barker, D.J. The developmental origins of adult disease. J. Am. Coll. Nutr. 2004, 23, 588S–595S.
- Goyal, D.; Limesand, S.W.; Goyal, R. Epigenetic responses and the developmental origins of health and disease. J. Endocrinol. 2019, 242, T105–T119.
- Holme, A.M.; Sitras, V. Developmental origin of health and disease—Evidence and time for action. Acta Obstet. Gynecol. Scand. 2020, 99, 961–962.
- Heindel, J.J.; Balbus, J.; Birnbaum, L.; Brune-Drisse, M.N.; Grandjean, P.; Gray, K.; Landrigan, P.J.; Sly, P.D.; Suk, W.A.; Cory Slechta, D.; et al. Developmental Origins of Health and Disease: Integrating Environmental Influences. Endocrinology 2015, 156, 3416–3421.
- Petronis, A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature 2010, 465, 721–727.
- Deichmann, U. Epigenetics: The origins and evolution of a fashionable topic. Dev. Biol. 2016, 416, 249–254.
- Chen, M.; Zhang, L. Epigenetic mechanisms in developmental programming of adult disease. Drug Discov. Today 2011, 16, 1007–1018.
- Banik, A.; Kandilya, D.; Ramya, S.; Stünkel, W.; Chong, Y.S.; Dheen, S.T. Maternal Factors that Induce Epigenetic Changes Contribute to Neurological Disorders in Offspring. Genes 2017, 8, 150.
- Barker, D.J.P. Sir Richard Doll Lecture. Developmental origins of chronic disease. Public Health 2012, 126, 185–189.
- Plagemann, A. Maternal diabetes and perinatal programming. Early Hum. Dev. 2011, 87, 743–747.
- Horta, B.L.; de Lima, N.P. Breastfeeding and Type 2 Diabetes: Systematic Review and Meta-Analysis. Curr. Diab. Rep. 2019, 19, 1.
- Camacho-Morales, A.; Caba, M.; García-Juárez, M.; Caba-Flores, M.D.; Viveros-Contreras, R.; Martínez-Valenzuela, C. Breastfeeding Contributes to Physiological Immune Programming in the Newborn. Front. Pediatr. 2021, 9, 744104.
- Ozkan, H.; Tuzun, F.; Taheri, S.; Korhan, P.; Akokay, P.; Yılmaz, O.; Duman, N.; Özer, E.; Tufan, E.; Kumral, A.; et al. Epigenetic Programming Through Breast Milk and Its Impact on Milk-Siblings Mating. Front. Genet. 2020, 11, 569232.
- Chutipongtanate, S.; Morrow, A.L.; Newburg, D.S. Human Milk Extracellular Vesicles: A Biological System with Clinical Implications. Cells 2022, 11, 2345.
- Tingö, L.; Ahlberg, E.; Johansson, L.; Pedersen, S.A.; Chawla, K.; Sætrom, P.; Cione, E.; Simpson, M.R. Non-Coding RNAs in Human Breast Milk: A Systematic Review. Front. Immunol. 2021, 12, 725323.
- Melnik, B.C.; Stremmel, W.; Weiskirchen, R.; John, S.M.; Schmitz, G. Exosome-Derived MicroRNAs of Human Milk and Their Effects on Infant Health and Development. Biomolecules 2021, 11, 851.
- Koh, K. Maternal breastfeeding and children’s cognitive development. Soc. Sci. Med. 2017, 187, 101–108.
- Wallenborn, J.T.; Levine, G.A.; Santos, A.C.D.; Grisi, S.; Brentani, A.; Fink, G. Breastfeeding, Physical Growth, and Cognitive Development. Pediatrics 2021, 147, e2020008029.
- Lewallen, L.P. Breastfeeding is important for cognitive development in term and preterm infants. Evid. Based Nurs. 2012, 15, 85–86.
- Horta, B.L.; de Mola, C.L.; Victora, C.G. Breastfeeding and intelligence: A systematic review and meta-analysis. Acta Paediatr. 2015, 104, 14–19.
- Kramer, M.S.; Aboud, F.; Mironova, E.; Vanilovich, I.; Platt, R.W.; Matush, L.; Igumnov, S.; Fombonne, E.; Bogdanovich, N.; Ducruet, T.; et al. Breastfeeding and child cognitive development: New evidence from a large randomized trial. Arch. Gen. Psychiatry 2008, 65, 578–584.
- Chetta, K.E.; Schulz, E.V.; Wagner, C.L. Outcomes improved with human milk intake in preterm and full-term infants. Semin. Perinatol. 2021, 45, 151384.
- Sullivan, G.; Vaher, K.; Blesa, M.; Galdi, P.; Stoye, D.Q.; Quigley, A.J.; Thrippleton, M.J.; Norrie, J.; Bastin, M.E.; Boardman, J.P. Breast Milk Exposure is Associated with Cortical Maturation in Preterm Infants. Ann. Neurol. 2022, 93, 591–603.
- Xu, J.; Shin, J.; McGee, M.; Unger, S.; Bando, N.; Sato, J.; Vandewouw, M.; Patel, Y.; Branson, H.M.; Paus, T.; et al. Intake of mother’s milk by very-low-birth-weight infants and variation in DNA methylation of genes involved in neurodevelopment at 5.5 years of age. Am. J. Clin. Nutr. 2022, 116, 1038–1048.
- Hu, Y.; Thaler, J.; Nieuwland, R. Extracellular Vesicles in Human Milk. Pharmaceuticals 2021, 14, 1050.
- He, S.; Liu, G.; Zhu, X. Human breast milk-derived exosomes may help maintain intestinal epithelial barrier integrity. Pediatr. Res. 2021, 90, 366–372.
- Shandilya, S.; Rani, P.; Onteru, S.K.; Singh, D. Small Interfering RNA in Milk Exosomes Is Resistant to Digestion and Crosses the Intestinal Barrier In Vitro. J. Agric. Food Chem. 2017, 65, 9506–9513.
- Melnik, B.C.; John, S.M.; Schmitz, G. Milk is not just food but most likely a genetic transfection system activating mTORC1 signaling for postnatal growth. Nutr. J. 2013, 12, 103.
- Chen, C.C.; Liu, L.; Ma, F.; Wong, C.W.; Guo, X.E.; Chacko, J.V.; Farhoodi, H.P.; Zhang, S.X.; Zimak, J.; Ségaliny, A.; et al. Elucidation of Exosome Migration across the Blood–Brain Barrier Model In Vitro. Cell. Mol. Bioeng. 2016, 9, 509–529.
- Green, D.; Dalmay, T.; Chapman, T. Microguards and micromessengers of the genome. Heredity 2016, 116, 125–134.
- Ying, S.-Y.; Chang, D.C.; Lin, S.-L. The microRNA (miRNA): Overview of the RNA genes that modulate gene function. Mol. Biotechnol. 2008, 38, 257–268.
- Carrillo-Lozano, E.; Sebastián-Valles, F.; Knott-Torcal, C. Circulating microRNAs in Breast Milk and Their Potential Impact on the Infant. Nutrients 2020, 12, 3066.
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51.
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297.
- Wang, Z. MicroRNA: A matter of life or death. World J. Biol. Chem. 2010, 1, 41–54.
- Hatmal, M.M.; Al-Hatamleh, M.A.I.; Olaimat, A.N.; Alshaer, W.; Hasan, H.; Albakri, K.A.; Alkhafaji, E.; Issa, N.N.; Al-Holy, M.A.; Abderrahman, S.M.; et al. Immunomodulatory Properties of Human Breast Milk: MicroRNA Contents and Potential Epigenetic Effects. Biomedicines 2022, 10, 1219.
- Aliperti, V.; Skonieczna, J.; Cerase, A. Long Non-Coding RNA (lncRNA) Roles in Cell Biology, Neurodevelopment and Neurological Disorders. Non Coding RNA 2021, 7, 36.
- Ng, S.Y.; Lin, L.; Soh, B.S.; Stanton, L.W. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013, 29, 461–468.
- Shi, C.; Zhang, L.; Qin, C. Long non-coding RNAs in brain development, synaptic biology, and Alzheimer’s disease. Brain Res. Bull. 2017, 132, 160–169.
- Lipovich, L.; Tarca, A.L.; Cai, J.; Jia, H.; Chugani, H.T.; Sterner, K.N.; Grossman, L.I.; Uddin, M.; Hof, P.R.; Sherwood, C.C.; et al. Developmental changes in the transcriptome of human cerebral cortex tissue: Long noncoding RNA transcripts. Cereb. Cortex 2014, 24, 1451–1459.
- Li, L.; Zhuang, Y.; Zhao, X.; Li, X. Long Non-coding RNA in Neuronal Development and Neurological Disorders. Front. Genet. 2018, 9, 744.
- Wilkinson, B.; Campbell, D.B. Contribution of long noncoding RNAs to autism spectrum disorder risk. Int. Rev. Neurobiol. 2013, 113, 35–59.
- Weissman, I.L. Translating stem and progenitor cell biology to the clinic: Barriers and opportunities. Science 2000, 287, 1442–1446.
- Kersin, S.G.; Ozek, E. Breast milk stem cells: Are they magic bullets in neonatology? Turk. Arch. Pediatr. 2021, 56, 187–191.
- Briere, C.E.; McGrath, J.M.; Jensen, T.; Matson, A.; Finck, C. Breast Milk Stem Cells: Current Science and Implications for Preterm Infants. Adv. Neonatal. Care 2016, 16, 410–419.
- Cregan, M.D.; Fan, Y.; Appelbee, A.; Brown, M.L.; Klopcic, B.; Koppen, J.; Mitoulas, L.R.; Piper, K.M.E.; Choolani, M.A.; Chong, Y.-S.; et al. Identification of nestin-positive putative mammary stem cells in human breastmilk. Cell Tissue Res. 2007, 329, 129–136.
- Briere, C.E.; Jensen, T.; McGrath, J.M.; Young, E.E.; Finck, C. Stem-Like Cell Characteristics from Breast Milk of Mothers with Preterm Infants as Compared to Mothers with Term Infants. Breastfeed. Med. 2017, 12, 174–179.
- Aydın, M.Ş.; Yiğit, E.N.; Vatandaşlar, E.; Erdoğan, E.; Öztürk, G. Transfer and Integration of Breast Milk Stem Cells to the Brain of Suckling Pups. Sci. Rep. 2018, 8, 14289.
- Belkozhayev, A.M.; Al-Yozbaki, M.; George, A.; Ye Niyazova, R.; Sharipov, K.O.; Byrne, L.J.; Wilson, C.M. Extracellular Vesicles, Stem Cells and the Role of miRNAs in Neurodegeneration. Curr. Neuropharmacol. 2022, 20, 1450–1478.
- Davis, C.; Savitz, S.I.; Satani, N. Mesenchymal Stem Cell Derived Extracellular Vesicles for Repairing the Neurovascular Unit after Ischemic Stroke. Cells 2021, 10, 767.
- Chang, Y.-H.; Wu, K.-C.; Harn, H.-J.; Lin, S.-Z.; Ding, D.-C. Exosomes and Stem Cells in Degenerative Disease Diagnosis and Therapy. Cell Transplant. 2018, 27, 349–363.
- Lendahl, U.; Zimmerman, L.B.; McKay, R.D.G. CNS stem cells express a new class of intermediate filament protein. Cell 1990, 60, 585–595.
- Hosseini, S.M.; Talaei-Khozani, T.; Sani, M.; Owrangi, B. Differentiation of human breast-milk stem cells to neural stem cells and neurons. Neurol. Res. Int. 2014, 2014, 807896.
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810.
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103.
- Gavin, A.; Ostovar, K.; Gavin, A.; Ostovar, K. Microbiological Characterization of Human Milk (1). J. Food Prot. 1977, 40, 614–616.
- Jones, C.A. Maternal transmission of infectious pathogens in breast milk. J. Paediatr. Child Health 2001, 37, 576–582.
- Martín, R.; Langa, S.; Reviriego, C.; Jimínez, E.; Marín, M.L.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr. 2003, 143, 754–758.
- Collado, M.; Delgado, S.; Maldonado, A.; Rodríguez, J. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett. Appl. Microbiol. 2009, 48, 523–528.
- Hunt, K.M.; Foster, J.A.; Forney, L.J.; Schütte, U.M.E.; Beck, D.L.; Abdo, Z.; Fox, L.K.; Williams, J.E.; McGuire, M.K.; McGuire, M.A. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE 2011, 6, e21313.
- Yi, D.Y.; Kim, S.Y. Human Breast Milk Composition and Function in Human Health: From Nutritional Components to Microbiome and MicroRNAs. Nutrients 2021, 13, 3094.
- Moossavi, S.; Sepehri, S.; Robertson, B.; Bode, L.; Goruk, S.; Field, C.J.; Lix, L.M.; de Souza, R.J.; Becker, A.B.; Mandhane, P.J.; et al. Composition and Variation of the Human Milk Microbiota Are Influenced by Maternal and Early-Life Factors. Cell Host Microbe 2019, 25, 324–335.e4.
- Zimmermann, P.; Curtis, N. Breast milk microbiota: A review of the factors that influence composition. J. Infect. 2020, 81, 17–47.
- Damaceno, Q.S.; Souza, J.P.; Nicoli, J.R.; Paula, R.L.; Assis, G.B.; Figueiredo, H.C.; Azevedo, V.; Martins, F.S. Evaluation of Potential Probiotics Isolated from Human Milk and Colostrum. Probiotics Antimicrob. Proteins 2017, 9, 371–379.
- Solís, G.; de los Reyes-Gavilan, C.G.; Fernández, N.; Margolles, A.; Gueimonde, M. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe 2010, 16, 307–310.
- Khodayar-Pardo, P.; Mira-Pascual, L.; Collado, M.C.; Martínez-Costa, C. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J. Perinatol. 2014, 34, 599–605.
- Dahaban, N.M.; Romli, M.F.; Roslan, N.R.; Kong, S.S.-S.; Cheah, F.-C. Bacteria in expressed breastmilk from mothers of premature infants and maternal hygienic status. Breastfeed. Med. 2013, 8, 422–423.
- Boix-Amorós, A.; Collado, M.C.; Mira, A. Relationship between Milk Microbiota, Bacterial Load, Macronutrients, and Human Cells during Lactation. Front. Microbiol. 2016, 7, 492.
- Hermansson, H.; Kumar, H.; Collado, M.C.; Salminen, S.; Isolauri, E.; Rautava, S. Breast Milk Microbiota Is Shaped by Mode of Delivery and Intrapartum Antibiotic Exposure. Front. Nutr. 2019, 6, 4.
- Simpson, M.R.; Avershina, E.; Storrø, O.; Johnsen, R.; Rudi, K.; Øien, T. Breastfeeding-associated microbiota in human milk following supplementation with Lactobacillus rhamnosus GG, Lactobacillus acidophilus La-5, and Bifidobacterium animalis ssp. lactis Bb-12. J. Dairy Sci. 2018, 101, 889–899.
- Gueimonde, M.; Laitinen, K.; Salminen, S.; Isolauri, E. Breast milk: A source of bifidobacteria for infant gut development and maturation? Neonatology 2007, 92, 64–66.
- Vuong, H.E. Intersections of the microbiome and early neurodevelopment. Int. Rev. Neurobiol. 2022, 167, 1–23.
- Lu, J.; Drobyshevsky, A.; Lu, L.; Yu, Y.; Caplan, M.S.; Claud, E.C. Microbiota from Preterm Infants Who Develop Necrotizing Enterocolitis Drives the Neurodevelopment Impairment in a Humanized Mouse Model. Microorganisms 2023, 11, 1131.
- Xia, J.; Claud, E.C. Gut Microbiome-Brain Axis as an Explanation for the Risk of Poor Neurodevelopment Out-come in Preterm Infants with Necrotizing Enterocolitis. Microorganisms 2023, 11, 1035.
- Indrio, F.; Martini, S.; Francavilla, R.; Corvaglia, L.; Cristofori, F.; Mastrolia, S.A.; Neu, J.; Rautava, S.; Spena, G.R.; Raimondi, F.; et al. Epigenetic Matters: The Link between Early Nutrition, Microbiome, and Long-term Health Development. Front. Pediatr. 2017, 5, 178.
- Tognini, P. Gut Microbiota: A Potential Regulator of Neurodevelopment. Front. Cell. Neurosci. 2017, 11, 25.
- Laue, H.E.; Coker, M.O.; Madan, J.C. The Developing Microbiome from Birth to 3 Years: The Gut-Brain Axis and Neurodevelopmental Outcomes. Front. Pediatr. 2022, 10, 815885.
- Ihekweazu, F.D.; Versalovic, J. Development of the Pediatric Gut Microbiome: Impact on Health and Disease. Am. J. Med Sci. 2018, 356, 413–423.
- Valicenti-McDermott, M.; McVicar, K.; Rapin, I.; Wershil, B.K.; Cohen, H.; Shinnar, S. Frequency of gastrointestinal symptoms in children with autistic spectrum disorders and association with family history of autoimmune disease. J. Dev. Behav. Pediatr. 2006, 27, S128–S136.
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013, 155, 1451–1463.
- Lu, J.; Claud, E.C. Connection between gut microbiome and brain development in preterm infants. Dev. Psychobiol. 2019, 61, 739–751.
