Ariadne’s thread on the path of our discovery of DNA and genetic code symmetries was our trinucleotide classification. Trinucleotides of each DNA genome and codons of the genetic code consist of four nitrogenous bases: two purines (adenine (A) and guanine (G)) and two pyrimidines (cytosine (C) and thymine (T) or uracil (U)). Thus, three of the bases are found in both DNA and the genetic code, whereas thymine is unique to DNA, and uracil is unique to the genetic code. A nucleotide is formed in the cell when the base attaches itself to the 1′ carbon of the sugar and phosphate attaches itself to the 5′ carbon of the same sugar the nucleotide takes its name from.
1. Introduction
Ten years ago, our main challenge was to find a solution as to how, in an autonomous system such as the DNA molecule, Chargaff’s second parity rule (CSPR) [
1], also called strand symmetry, can function: Why is the relative frequency of some trinucleotides almost identical to the relative frequency of their reverse complement in the same DNA strand of about 100 kb or more? What is common in, for example, ATG (direct) and CAT (reverse complement), or CAA (direct) and TTG (reverse complement)? What has kept such symmetry unchanged for all DNA prokaryotes and eukaryotes during all of evolution?
The fundamental role of symmetry in a biological system is to decrease disorder (entropy) and to preserve the integrity of the system. Many scientists have investigated the importance of symmetries in biological processes, especially after the discovery of the DNA molecule and the genetic code as basic structures related to life on Earth. The general question is whether symmetries reflect some fundamental “laws” of genome evolution or whether they are a type of statistical pattern [
2]. The idea that natural laws are associated with some symmetry is widespread, but the symbiosis of mathematics and natural laws was not fully understood [
3,
4]. Earlier, in 1918, Emmy Nöther proved her famous theorem by relating symmetry in time to the energy conservation law [
5]. As pointed out by Gross [
6], Einstein’s great advance was to put the symmetries as a dominant concept in the fundamental laws of physics, to regard the symmetry principle as the primary feature of nature: the symmetry principles dictate the form of the laws of nature. Einstein’s paradigm implies, in general, a broader view on the problem of the evolution of natural laws. For example, the law of energy conservation is a natural consequence of the existence of time symmetry, and not of some kind of evolution. Analogically, according to Einstein’s paradigm, the Supersymmetry Genetic Code (SSyGC), which is unchangeable during whole evolution, could be considered as a natural consequence of physicochemical symmetries with the corresponding mirror symmetry, and not being generated by an evolutionary process.
Jacques Monod attached great significance to symmetry in biology, which must not be understood in purely geometrical connotations, but rather in a much wider sense, identical to that of order within a structure [
7].
In 1943, Schrödinger proposed in his lecture at Trinity College in Dublin that hereditary material must take the form of an “aperiodic crystal”, implying the presence of symmetries in the structure of DNA. Ever since Nirenberg’s discovery in 1961 (codons code for individual amino acids), scientists searched for symmetries within genetic code. Up to the present discovery of the SSyGC, complete symmetry in the genetic code has not been found, leaving a doubt as to whether the symmetrical nature as the “protector” of order even exists.
2. Classification of Trinucleotides/Codons
Ariadne’s thread on the path of our discovery of DNA and genetic code symmetries was our trinucleotide classification [
8,
9,
10]. Trinucleotides of each DNA genome and codons of the genetic code consist of four nitrogenous bases: two purines (adenine (A) and guanine (G)) and two pyrimidines (cytosine (C) and thymine (T) or uracil (U)). Thus, three of the bases are found in both DNA and the genetic code, whereas thymine is unique to DNA, and uracil is unique to the genetic code. A nucleotide is formed in the cell when the base attaches itself to the 1′ carbon of the sugar and phosphate attaches itself to the 5′ carbon of the same sugar the nucleotide takes its name from.
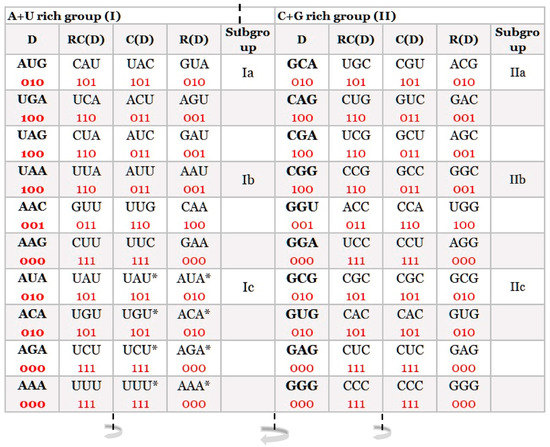
At first sight, a simple distribution of trinucleotides on A + T rich and C + G rich showed an important basic structural organization of our DNA classification in the form of 20 quadruplets: 10 A + T rich and 10 C + G rich (Figure 1). Each trinucleotide is very important because they represent the basic structure of the DNA molecule, while the genetic code represents the code in the form of codons for 20 natural amino acids, which are the fundamental structure for the protein synthesis of living species.
Figure 1. Our quadruplet classification of 64 codons (with U—uracil) for the genetic code, or trinucleotides (with T—thymine instead of uracil) for RNA and DNA genomes. Each quadruplet is unique and consists of four specific codons or trinucleotides denoted as direct D, reverse complement from direct RC(D), complement from direct C(D), and reverse from direct R(D). Ten A + U rich (group I) and ten C + G rich (group II) quadruplets are organized in three subgroups. Ia consisting of nonsymmetrical codons/trinucleotides containing three different nucleotides, Ib consisting of nonsymmetrical codons/trinucleotides containing two different nucleotides, and Ic consisting of symmetrical codons/trinucleotides that contain duplicated codons/trinucleotides labelled with an asterisk (D = RC, C = R). The first four A + U rich quadruplets were generated with start/stop signals: AUG, UGA, UAG, and UAA. The C + G rich trinucleotides correspond to the purine–purine and pyrimidine–pyrimidine transformation of A + U rich codons/trinucleotides. Three symmetries are present in our codon/trinucleotide classification: (1) purine–pyrimidine symmetries in each quadruplet, (2) purine–pyrimidine symmetries within and between A + U rich and C + G rich quadruplets in the same row of the classification, and (3) mirror symmetry between the direct-reverse and complement-reverse complement in the same quadruplet. Mirror symmetry is also present between purines and pyrimidines of the whole A + T rich group and C + G rich group of codons/trinucleotides. For clarity, the white and grey rows are alternating, to emphasize pairs of A + T rich and C + G rich codons. 0, purine; 1, pyrimidine. It is irrelevant which codon/trinucleotide in the quadruplet is direct, because the other three are accordingly adapted: mirror symmetry. From work by Marija Rosandić and Vladimir Paar [
11], published by Elsevier and reproduced with the permission of the publisher.
From the classification of trinucleotides/codons, a symmetrical relationship between purines and pyrimidines in each quadruplet is the same between direct (D) and reverse (R), as well as between complement (C) and reverse complement (RC). Purines marked as “0” and pyrimidines as “1” for trinucleotides and codons give eight possible combinations from which the first two are in a D↔C relationship, and the other two are in a D↔R relationship: [000 ↔ 111, 010 ↔ 101], [100 ↔ 001, 011 ↔ 110]. The point is that all four members of each quadruplet are specific, and it is free choice which is considered as direct (D). The others are adjusted according to Watson–Crick pairing for complement, reverse, and reverse complement functions (Figure 1).
This entry is adapted from the peer-reviewed paper 10.3390/ijms241512029