Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Despite the declining trend of Helicobacter pylori (H. pylori) prevalence around the globe, ongoing efforts are still needed to optimize current and future regimens in view of the increasing antibiotic resistance. The resistance of H. pylori to different antibiotics is caused by different molecular mechanisms, and advancements in sequencing technology have come a far way in broadening our understanding and in facilitating the testing of antibiotic susceptibility to H. pylori.
- H. pylori
- vonoprazan
- bismuth
1. Introduction
Helicobacter pylori (H. pylori) infects approximately 40% of individuals worldwide [1] and is involved in various gastrointestinal diseases such as chronic gastritis, peptic ulcer disease, upper gastrointestinal bleeding [2], gastric cancer (including gastric adenocarcinoma and gastric MALT lymphoma) [3][4][5] and extraintestinal manifestations [6]. All patients infected with H. pylori should be treated regardless of clinical manifestations [4]. Eradication of H. pylori can restore the normal gastric mucosa [7] and was shown to effectively reduce the development and recurrence of peptic ulcers [8][9], as well as the incidence of gastric cancer [10][11].
Various treatment regimens for H. pylori have been developed and their use vary across different geographical regions largely based on local antibiotic resistance patterns. Currently, an acceptable H. pylori treatment regimen is defined as one that achieves at least a 90% cure rate [12], though it has been suggested that an optimized regimen should reliably achieve ≥95% cure rates [13]. Treatment regimens currently recommended by various international guidelines are of empirical nature [4][14][15][16]. In light of a substantial decline in efficacy to levels below 80–85% [12][17], triple therapy consisting of a proton pump inhibitor (PPI), clarithromycin, and amoxicillin or metronidazole is usually not recommended as an empirical first-line therapy for H. pylori infection, except in areas with a known clarithromycin resistance rate of <15% [4][14][15]. Instead, bismuth-based quadruple therapy, typically consisting of bismuth, PPI, metronidazole and tetracycline, is increasingly recommended as its replacement for first-line treatment [4][14][15] and has been shown to achieve over 90% success rate [18][19]. Bismuth-based quadruple therapy can also be used as a second-line treatment. Another option for second-line treatment is a triple therapy consisting of a PPI, amoxicillin and levofloxacin, but this too faces the problem of decreasing efficacy because of rising resistance to levofloxacin [20]. It is therefore important to deepen our understanding of the mechanisms of antibiotic resistance in H. pylori through the use of various technologies, such as next-generational sequencing (NGS). Antibiotic resistance patterns should be regularly monitored and shared with physicians to guide clinical decisions regarding the antibiotic combinations used for eradication therapy.
2. Antibiotic Resistance in Helicobacter pylori
Primary antibiotic resistance in H. pylori treatment can be defined as resistance in patients who have not started eradication therapy, whereas secondary antibiotic resistance occurs in patients who have previously undergone at least one unsuccessful eradication attempt [21]. Globally, primary and secondary resistance rates of clarithromycin, levofloxacin and metronidazole were all over 15% in all World Health Organization (WHO) regions [21]. The underlying mechanism behind antibiotic resistance by H. pylori is often due to genetic mutations that may inhibit the intracellular activation of antibiotics or change the drug target site altogether [22], and these mutations are mostly encoded chromosomally rather than in extrachromosomal elements [23][24][25]. The specifics of such genetic changes depend on the class of antibiotics (Figure 1) (Table 1). Apart from genetic sequence mutations, there are other possible mechanisms such as efflux pumps through the upregulation or downregulation of hefABC genes, or through biofilm formation, which can obstruct drug penetration [22] (Figure 1).
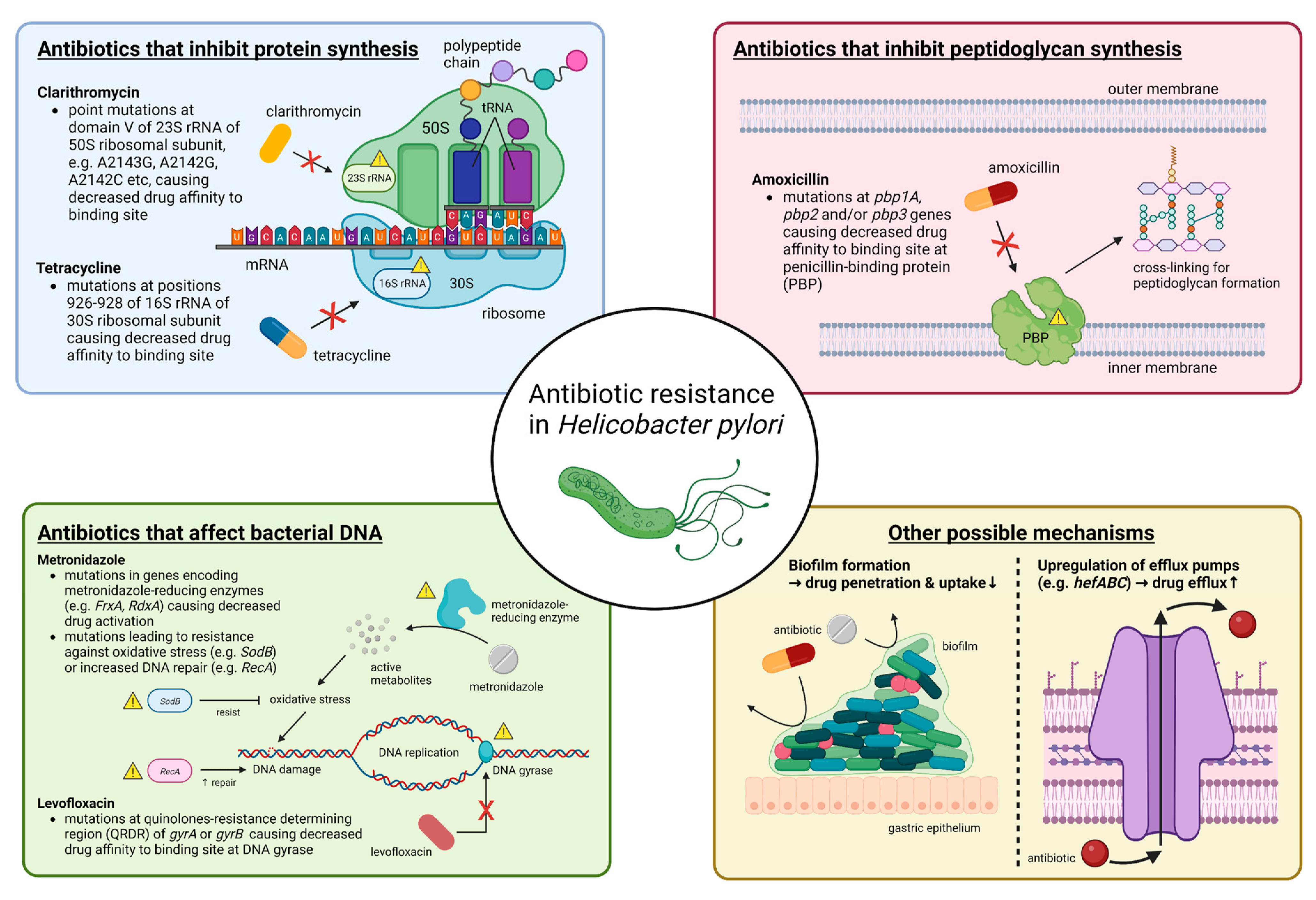
Figure 1. Summary of molecular mechanisms of antibiotic resistance in H. pylori. In general, antibiotic resistance in H. pylori arises because of mutations that decrease drug affinity to its binding site. Examples include mutations in 23S rRNA for clarithromycin, in pbp1A for amoxicillin, in gyrA or gyrB for levofloxacin and in 16S rRNA for tetracycline. Metronidazole is unique in that it is a prodrug and works by producing active metabolites that can damage bacterial DNA after reductive activation. Mutations causing metronidazole resistance have a much more diverse repertoire, and more well-known ones include those in genes encoding for metronidazole-reducing enzymes (e.g., FrxA and RdxA) causing decreased drug activation, as well as those that lead to resistance to oxidative stress caused by metronidazole or increased DNA repair. Other possible mechanisms that may apply to most antibiotics include increased drug efflux caused by upregulation of efflux pump genes (e.g., hefABC) or biofilm formation, which decrease drug penetration. Abbreviations: rRNA, ribosomal RNA; mRNA, messenger RNA; tRNA, transfer RNA; DNA, deoxyribonucleic acid.
Table 1. Molecular basis of resistance against antibiotics commonly used in Helicobacter pylori eradication therapies.
| Genes Involved | Resistance Mechanisms | |
|---|---|---|
| Clarithromycin | Domain V of 23S rRNA (most prevalent: A2143G, A2142G, A2142C) [22][26][27][28] | Decrease drug affinity to its binding site |
| infB (translation initiation factor IF-2), Rpl22 (Ribosomal protein L22) [29][30][31] |
Putative effects on 23S rRNA (the drug target) * | |
| Novel gene candidate: fliJ (flagellar export protein): Gln31Arg [32] |
Putative effects on bacterial chemotaxis and flagellar motility * | |
| Metronidazole | RdxA (oxygen-insensitive NAD(P)H nitroreductase), FrxA (NAD(P)H flavin nitroreductase), |
Mutations in metronidazole-reducing enzymes coding genes causing decreased drug activation |
| FdxA (ferredoxin), FdxB (ferredoxin-like protein), FldA (flavodoxin) [22] |
Mutations in putative metronidazole-reducing enzymes coding genes causing decreased drug activation | |
| SodB (superoxide dismutase), Fur (ferric uptake regulator), MdaB (modulator of drug activity B) [22] |
Resistance against oxidative stress brought by metronidazole | |
| RecA [22] | Upregulation of RecA causing enhanced DNA repair against damages brought by metronidazole | |
| Ribf (riboflavin biosynthesis protein), Omp11 [22] |
Putative association with metronidazole resistance * | |
| hefA (efflux pump) [33][34] | Increased drug efflux causing below lethal intracellular concentrations | |
| Levofloxacin | gyrA (DNA gyrase subunit A): mainly at codons N87 or D91, gyrB (DNA gyrase subunit B) [22][35] |
Mutations in quinolones resistance-determining region (QRDR) causing decreased drug affinity to its binding site |
| Novel gene candidates: fliJ (flagellar export protein): Gln31Arg [32] cheA (histidine kinase): Ser176Thr/Ala [32] |
Putative effects on bacterial chemotaxis and flagellar motility * | |
| Amoxicillin | pbp1A (penicillin-binding protein 1A) [36][37] pbp2 (penicillin-binding protein 2) [38] pbp3 (penicillin-binding protein 3) [38] |
Mutations altering penicillin-binding motifs SXXK, SXN and KTG, causing decreased drug affinity to its binding site |
| hopB, hopC (porins), hefC (efflux pump) [39][40] |
Increased drug efflux causing below lethal intracellular concentrations | |
| Tetracycline | 16S rRNA positions 926–928 [41][42][43] | Decrease drug affinity to its binding site |
| hefABC (efflux pump) [44] | Increased drug efflux causing below lethal intracellular concentrations |
* Hypothetical mechanism.
2.1. Resistance to Clarithromycin
Clarithromycin resistance in H. pylori is most commonly due to point mutations in domain V of 23S rRNA, especially A2143G, A2142G and A2142C [22][26][27][28], with A2143G causing the lowest eradication rate [45][46]. Other mutations have also been observed in [47][48][49] and outside of domain V [50], as well as in genes outside of 23S rRNA, such as infB and rpl22, which putatively affect 23S rRNA [29][30][31]. Increasing clarithromycin resistance was responsible for the failure of clarithromycin-containing eradication therapies [4][20][51]. The primary clarithromycin resistance rate was below 15% only in the WHO regions of the Americas and Southeast Asia, and when secondary resistance was considered, all WHO regions had >15% resistance rate [21], which has exceeded the suggested threshold at which clarithromycin triple therapy could be used as an empirical first-line treatment [52]. Studies found that primary clarithromycin resistance rates were 17–27% in Asia [53][54], 25% in treatment-naïve patients in Europe [55], 17.6–31.5% in the US [56][57] and 29.2% in Africa [58].
Of note, resistance caused by prior usage of clarithromycin or other macrolides for other diseases in patients who had never taken clarithromycin-based eradication therapy may be considered as secondary resistance [59][60]. Information on prior macrolide use, though difficult to obtain in real practice [61], should be collected whenever possible as cross resistance can occur within the same family of antibiotics [20]. Studies showed that macrolide exposure even in the past 10 to 14 years still correlated with H. pylori eradication failure [60][61][62][63][64] and was an independent risk factor for H. pylori clarithromycin resistance [60]. One such study found that those who previously took macrolides for >2 weeks had a significantly higher failure rate of eradication than those who took macrolides for ≤2 weeks (44.8% vs. 29.3%, p = 0.047), suggesting a duration effect of prior macrolide use on the success rate of eradication therapy [62].
2.2. Resistance to Metronidazole
The mutational changes involved in metronidazole resistance have a much more diverse repertoire and the genotype–phenotype correlation is often much more complex [4][22]. More well-known ones included RdxA and FrxA mutations, which were metronidazole-reducing enzymes and resulted in reduced drug activation [22]. Other putative mutations might result in increased efficiency of deoxyribonucleic acid (DNA) repair or oxidative stress response [22] and upregulated efflux [33][34]. It is possible that metronidazole resistance is caused by the cumulative effects of multiple pathways rather than a single mechanism, though until now, there has been no study that comprehensively investigated these mechanisms in the same clinical isolates [22].
The metronidazole resistance rate is generally much higher than that of clarithromycin, in part because of its widespread use to treat parasitic infections, urinary tract infections and gastrointestinal infections by anaerobes [65]. The primary and secondary resistance rates of metronidazole were over 15% in all WHO regions, with the highest rates of 56% and 65%, respectively, observed in the eastern Mediterranean region [21]. An increasing trend of metronidazole resistance has been observed in most of the WHO regions [21]. Studies have found that primary metronidazole resistance was 44% in the Asia-Pacific region [53], 30–40% in Europe [55][66], 42–43% in the US [56][57] and 48.7% in Africa [67]. A notable issue was the dual resistance to clarithromycin and metronidazole, which had a prevalence rate of around 8–15% in Europe [21][55][66], 6–11% in Asia [21] and 3–11% in the Americas [21][56][57]. Such dual resistance greatly reduces the efficacy of non-bismuth quadruple therapy [68][69]. In contrast to clarithromycin, in vitro metronidazole resistance can be overcome by increasing the dose, frequency and duration of therapy [52][70][71][72].
2.3. Resistance to Levofloxacin
As H. pylori does not naturally possess the genes for topoisomerase IV, levofloxacin resistance in H. pylori mainly arises from point mutations in the quinolone resistance-determining region (QRDR) of the gyrA or, to a lesser extent, gyrB gene, which code for DNA gyrase subunits A and B, respectively [22][35]. Mutations at gyrA mainly occur at codons N87 or D91 [73][74], while E463 mutation has been observed at gyrB [75][76].
Primary resistance against levofloxacin exceeded 15% in all WHO regions except Europe, while secondary resistance exceeded 15% in all regions with a significant rising trend observed in the western Pacific WHO region [21]. Other studies found that the primary levofloxacin resistance rates were 18% in the Asia-Pacific region [53], 15–20% in Europe [55][66], 37.6% in the US [57] and 17.4% in Africa [58]. Multiple resistance, commonly as dual or triple resistance to clarithromycin and/or metronidazole, is also an area of concern, with rates up to 6–10% in Europe [55][66].
2.4. Resistance to Amoxicillin
Resistance to amoxicillin in H. pylori are mainly caused by mutations in the pbp1A gene [36][37] that altered the penicillin-binding motifs SXXK, SXN and KTG, hence reducing the affinity of amoxicillin to PBP [22][26]. Enhancing effects can be contributed to by mutations in pbp2 and pbp3, and if mutations in these three genes occur simultaneously, resistance could increase by over 200-fold [38]. Mutations of porin genes (e.g., hopB and hopC) or efflux pump-coding genes (e.g., hefC) can contribute to amoxicillin resistance in H. pylori as well [39][40].
Amoxicillin resistance in H. pylori is generally lower than other antibiotics. Primary and secondary amoxicillin resistance rates were previously found to be below 15% in all WHO regions [21]. Low resistance was generally found in more-developed regions, such as 6.4% in the US [56], 3% in the Asia-Pacific region [53] and 0.2% in Europe [66]. In Africa, the resistance rate could be as high as 72.6% [58], likely caused by the over abuse of amoxicillin because of low cost [77].
2.5. Resistance to Tetracycline
Tetracycline resistance in H. pylori is mainly caused by mutations at positions 926–928 of the 16S rRNA [41][42][43] that can lead to reduced drug affinity as they are located at the primary binding site [78]. Among these mutations, triple base-pair mutations (e.g., AGA → TTC) conferred a higher level of tetracycline resistance than single or double base-pair mutations [79]. Other possible mechanisms included efflux pump mechanisms that might be mediated by HefABC [44] or by proton motive force (PMF)-dependent mechanisms [80], which might explain resistance in H. pylori strains without 16S rRNA mutations.
Similar to amoxicillin, resistance against tetracycline is generally low around the world, with primary and secondary resistance below 10% in all WHO regions [21]. The overall resistance rate of tetracycline was 4% in the Asia-Pacific region [53], below 1% in Europe [55][66] and below 3% in the US [56][57]. In Africa, however, misuse of tetracyclines caused the resistance rate to be much higher at 49.8% [58], which might cause reduced efficacy of bismuth quadruple therapy locally [52].
This entry is adapted from the peer-reviewed paper 10.3390/ijms241411708
References
- Li, Y.; Choi, H.; Leung, K.; Jiang, F.; Graham, D.Y.; Leung, W.K. Global prevalence of Helicobacter pylori infection between 1980 and 2022: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2023, 8, 553–564.
- Guo, C.G.; Cheung, K.S.; Zhang, F.; Chan, E.W.; Chen, L.; Wong, I.C.K.; Leung, W.K. Delay in Retreatment of Helicobacter pylori Infection Increases Risk of Upper Gastrointestinal Bleeding. Clin. Gastroenterol. Hepatol. 2021, 19, 314–322.
- Malfertheiner, P.; Megraud, F.; O’Morain, C.; Bazzoli, F.; El-Omar, E.; Graham, D.; Hunt, R.; Rokkas, T.; Vakil, N.; Kuipers, E.J. Current concepts in the management of Helicobacter pylori infection: The Maastricht III Consensus Report. Gut 2007, 56, 772–781.
- Malfertheiner, P.; Megraud, F.; Rokkas, T.; Gisbert, J.P.; Liou, J.M.; Schulz, C.; Gasbarrini, A.; Hunt, R.H.; Leja, M.; O’Morain, C.; et al. Management of Helicobacter pylori infection: The Maastricht VI/Florence consensus report. Gut 2022, 71, 1724–1762.
- Cheung, K.S.; Leung, W.K. Risk of gastric cancer development after eradication of Helicobacter pylori. World J. Gastrointest. Oncol. 2018, 10, 115–123.
- Wong, F.; Rayner-Hartley, E.; Byrne, M.F. Extraintestinal manifestations of Helicobacter pylori: A concise review. World J. Gastroenterol. 2014, 20, 11950–11961.
- Tepes, B.; Kavcic, B.; Zaletel, L.K.; Gubina, M.; Ihan, A.; Poljak, M.; Krizman, I. Two- to four-year histological follow-up of gastric mucosa after Helicobacter pylori eradication. J. Pathol. 1999, 188, 24–29.
- Boltin, D.; Niv, Y.; Schutte, K.; Schulz, C. Review: Helicobacter pylori and non-malignant upper gastrointestinal diseases. Helicobacter 2019, 24 (Suppl. 1), e12637.
- Hentschel, E.; Brandstatter, G.; Dragosics, B.; Hirschl, A.M.; Nemec, H.; Schutze, K.; Taufer, M.; Wurzer, H. Effect of ranitidine and amoxicillin plus metronidazole on the eradication of Helicobacter pylori and the recurrence of duodenal ulcer. N. Engl. J. Med. 1993, 328, 308–312.
- Lee, Y.C.; Chiang, T.H.; Chou, C.K.; Tu, Y.K.; Liao, W.C.; Wu, M.S.; Graham, D.Y. Association Between Helicobacter pylori Eradication and Gastric Cancer Incidence: A Systematic Review and Meta-analysis. Gastroenterology 2016, 150, 1113–1124.
- Chiang, T.H.; Chang, W.J.; Chen, S.L.; Yen, A.M.; Fann, J.C.; Chiu, S.Y.; Chen, Y.R.; Chuang, S.L.; Shieh, C.F.; Liu, C.Y.; et al. Mass eradication of Helicobacter pylori to reduce gastric cancer incidence and mortality: A long-term cohort study on Matsu Islands. Gut 2021, 70, 243–250.
- Nyssen, O.P.; Bordin, D.; Tepes, B.; Perez-Aisa, A.; Vaira, D.; Caldas, M.; Bujanda, L.; Castro-Fernandez, M.; Lerang, F.; Leja, M.; et al. European Registry on Helicobacter pylori management (Hp-EuReg): Patterns and trends in first-line empirical eradication prescription and outcomes of 5 years and 21,533 patients. Gut 2021, 70, 40–54.
- Graham, D.Y.; Liou, J.M. Primer for Development of Guidelines for Helicobacter pylori Therapy Using Antimicrobial Stewardship. Clin. Gastroenterol. Hepatol. 2022, 20, 973–983.
- Chey, W.D.; Leontiadis, G.I.; Howden, C.W.; Moss, S.F. ACG Clinical Guideline: Treatment of Helicobacter pylori Infection. Am. J. Gastroenterol. 2017, 112, 212–239.
- Fallone, C.A.; Chiba, N.; van Zanten, S.V.; Fischbach, L.; Gisbert, J.P.; Hunt, R.H.; Jones, N.L.; Render, C.; Leontiadis, G.I.; Moayyedi, P.; et al. The Toronto Consensus for the Treatment of Helicobacter pylori Infection in Adults. Gastroenterology 2016, 151, 51–69.
- Katelaris, P.; Hunt, R.; Bazzoli, F.; Cohen, H.; Fock, K.M.; Gemilyan, M.; Malfertheiner, P.; Megraud, F.; Piscoya, A.; Quach, D.; et al. Helicobacter pylori World Gastroenterology Organization Global Guideline. J. Clin. Gastroenterol. 2023, 57, 111–126.
- Graham, D.Y.; Fischbach, L. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut 2010, 59, 1143–1153.
- Sun, Q.; Liang, X.; Zheng, Q.; Liu, W.; Xiao, S.; Gu, W.; Lu, H. High efficacy of 14-day triple therapy-based, bismuth-containing quadruple therapy for initial Helicobacter pylori eradication. Helicobacter 2010, 15, 233–238.
- Hsu, P.I.; Chen, K.Y.; Tai, W.C.; Yang, J.C.; Tsay, F.W.; Liu, Y.H.; Chen, C.L.; Lee, C.L.; Yeh, H.Z.; Kuo, C.H.; et al. Hybrid, High-Dose Dual and Bismuth Quadruple Therapies for First-Line Treatment of Helicobacter pylori Infection in Taiwan: A Multicenter, Open-Label, Randomized Trial. Am. J. Gastroenterol. 2023, 118, 1184–1195.
- Malfertheiner, P.; Megraud, F.; O’Morain, C.A.; Atherton, J.; Axon, A.T.; Bazzoli, F.; Gensini, G.F.; Gisbert, J.P.; Graham, D.Y.; Rokkas, T.; et al. Management of Helicobacter pylori infection--the Maastricht IV/Florence Consensus Report. Gut 2012, 61, 646–664.
- Savoldi, A.; Carrara, E.; Graham, D.Y.; Conti, M.; Tacconelli, E. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Systematic Review and Meta-analysis in World Health Organization Regions. Gastroenterology 2018, 155, 1372–1382.
- Tshibangu-Kabamba, E.; Yamaoka, Y. Helicobacter pylori infection and antibiotic resistance-from biology to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 613–629.
- Tuan, V.P.; Narith, D.; Tshibangu-Kabamba, E.; Dung, H.D.Q.; Viet, P.T.; Sokomoth, S.; Binh, T.T.; Sokhem, S.; Tri, T.D.; Ngov, S.; et al. A Next-Generation Sequencing-Based Approach to Identify Genetic Determinants of Antibiotic Resistance in Cambodian Helicobacter pylori Clinical Isolates. J. Clin. Med. 2019, 8, 858.
- Lauener, F.N.; Imkamp, F.; Lehours, P.; Buissonniere, A.; Benejat, L.; Zbinden, R.; Keller, P.M.; Wagner, K. Genetic Determinants and Prediction of Antibiotic Resistance Phenotypes in Helicobacter pylori. J. Clin. Med. 2019, 8, 53.
- Tshibangu-Kabamba, E.; Ngoma-Kisoko, P.J.; Tuan, V.P.; Matsumoto, T.; Akada, J.; Kido, Y.; Tshimpi-Wola, A.; Tshiamala-Kashala, P.; Ahuka-Mundeke, S.; Ngoy, D.M.; et al. Next-Generation Sequencing of the Whole Bacterial Genome for Tracking Molecular Insight into the Broad-Spectrum Antimicrobial Resistance of Helicobacter pylori Clinical Isolates from the Democratic Republic of Congo. Microorganisms 2020, 8, 887.
- Hu, Y.; Zhang, M.; Lu, B.; Dai, J. Helicobacter pylori and Antibiotic Resistance, A Continuing and Intractable Problem. Helicobacter 2016, 21, 349–363.
- Gong, Y.; Yuan, Y. Resistance mechanisms of Helicobacter pylori and its dual target precise therapy. Crit. Rev. Microbiol. 2018, 44, 371–392.
- Versalovic, J.; Shortridge, D.; Kibler, K.; Griffy, M.V.; Beyer, J.; Flamm, R.K.; Tanaka, S.K.; Graham, D.Y.; Go, M.F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 1996, 40, 477–480.
- Binh, T.T.; Shiota, S.; Suzuki, R.; Matsuda, M.; Trang, T.T.; Kwon, D.H.; Iwatani, S.; Yamaoka, Y. Discovery of novel mutations for clarithromycin resistance in Helicobacter pylori by using next-generation sequencing. J. Antimicrob. Chemother. 2014, 69, 1796–1803.
- Miftahussurur, M.; Shrestha, P.K.; Subsomwong, P.; Sharma, R.P.; Yamaoka, Y. Emerging Helicobacter pylori levofloxacin resistance and novel genetic mutation in Nepal. BMC Microbiol. 2016, 16, 256.
- Miftahussurur, M.; Syam, A.F.; Nusi, I.A.; Makmun, D.; Waskito, L.A.; Zein, L.H.; Akil, F.; Uwan, W.B.; Simanjuntak, D.; Wibawa, I.D.; et al. Surveillance of Helicobacter pylori Antibiotic Susceptibility in Indonesia: Different Resistance Types among Regions and with Novel Genetic Mutations. PLoS ONE 2016, 11, e0166199.
- Lyu, T.; Cheung, K.S.; Deng, Z.; Ni, L.; Chen, C.; Wu, J.; Leung, W.K.; Seto, W.K. Whole genome sequencing reveals novel genetic mutations of Helicobacter pylori associating with resistance to clarithromycin and levofloxacin. Helicobacter 2023, 28, e12972.
- Lee, S.M.; Kim, N.; Kwon, Y.H.; Nam, R.H.; Kim, J.M.; Park, J.Y.; Lee, Y.S.; Lee, D.H. rdxA, frxA, and efflux pump in metronidazole-resistant Helicobacter pylori: Their relation to clinical outcomes. J. Gastroenterol. Hepatol. 2018, 33, 681–688.
- van Amsterdam, K.; Bart, A.; van der Ende, A. A Helicobacter pylori TolC efflux pump confers resistance to metronidazole. Antimicrob. Agents Chemother. 2005, 49, 1477–1482.
- Puah, S.M.; Goh, K.L.; Ng, H.K.; Chua, K.H. Current status of Helicobacter pylori resistance to Clarithromycin and Levofloxacin in Malaysia-findings from a molecular based study. PeerJ 2021, 9, e11518.
- Gerrits, M.M.; Godoy, A.P.; Kuipers, E.J.; Ribeiro, M.L.; Stoof, J.; Mendonca, S.; van Vliet, A.H.; Pedrazzoli, J., Jr.; Kusters, J.G. Multiple mutations in or adjacent to the conserved penicillin-binding protein motifs of the penicillin-binding protein 1A confer amoxicillin resistance to Helicobacter pylori. Helicobacter 2006, 11, 181–187.
- Okamoto, T.; Yoshiyama, H.; Nakazawa, T.; Park, I.D.; Chang, M.W.; Yanai, H.; Okita, K.; Shirai, M. A change in PBP1 is involved in amoxicillin resistance of clinical isolates of Helicobacter pylori. J. Antimicrob. Chemother. 2002, 50, 849–856.
- Rimbara, E.; Noguchi, N.; Kawai, T.; Sasatsu, M. Mutations in penicillin-binding proteins 1, 2 and 3 are responsible for amoxicillin resistance in Helicobacter pylori. J. Antimicrob. Chemother. 2008, 61, 995–998.
- Co, E.M.; Schiller, N.L. Resistance mechanisms in an in vitro-selected amoxicillin-resistant strain of Helicobacter pylori. Antimicrob. Agents Chemother. 2006, 50, 4174–4176.
- Qureshi, N.N.; Gallaher, B.; Schiller, N.L. Evolution of amoxicillin resistance of Helicobacter pylori in vitro: Characterization of resistance mechanisms. Microb. Drug. Resist. 2014, 20, 509–516.
- Dailidiene, D.; Bertoli, M.T.; Miciuleviciene, J.; Mukhopadhyay, A.K.; Dailide, G.; Pascasio, M.A.; Kupcinskas, L.; Berg, D.E. Emergence of tetracycline resistance in Helicobacter pylori: Multiple mutational changes in 16S ribosomal DNA and other genetic loci. Antimicrob. Agents Chemother. 2002, 46, 3940–3946.
- Gerrits, M.M.; de Zoete, M.R.; Arents, N.L.; Kuipers, E.J.; Kusters, J.G. 16S rRNA mutation-mediated tetracycline resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 2002, 46, 2996–3000.
- Trieber, C.A.; Taylor, D.E. Mutations in the 16S rRNA genes of Helicobacter pylori mediate resistance to tetracycline. J. Bacteriol. 2002, 184, 2131–2140.
- Kutschke, A.; de Jonge, B.L. Compound efflux in Helicobacter pylori. Antimicrob. Agents Chemother. 2005, 49, 3009–3010.
- Versalovic, J.; Osato, M.S.; Spakovsky, K.; Dore, M.P.; Reddy, R.; Stone, G.G.; Shortridge, D.; Flamm, R.K.; Tanaka, S.K.; Graham, D.Y. Point mutations in the 23S rRNA gene of Helicobacter pylori associated with different levels of clarithromycin resistance. J. Antimicrob. Chemother. 1997, 40, 283–286.
- Alfaresi, M.S.; Eltom, A.A.; Alshaikh, S.S.; Buchel, E.H.; Saeed, L.S.; Aljenaibi, M.M. Molecular prevalence of point mutations conferring resistance to clarithromycin in Helicobacter pylori in the United Arab Emirates. Saudi Med. J. 2005, 26, 763–766.
- Khan, R.; Nahar, S.; Sultana, J.; Ahmad, M.M.; Rahman, M. T2182C mutation in 23S rRNA is associated with clarithromycin resistance in Helicobacter pylori isolates obtained in Bangladesh. Antimicrob. Agents Chemother. 2004, 48, 3567–3569.
- Toracchio, S.; Aceto, G.M.; Mariani-Costantini, R.; Battista, P.; Marzio, L. Identification of a novel mutation affecting domain V of the 23S rRNA gene in Helicobacter pylori. Helicobacter 2004, 9, 396–399.
- Garrido, L.; Toledo, H. Novel genotypes in Helicobacter pylori involving domain V of the 23S rRNA gene. Helicobacter 2007, 12, 505–509.
- Hao, Q.; Li, Y.; Zhang, Z.J.; Liu, Y.; Gao, H. New mutation points in 23S rRNA gene associated with Helicobacter pylori resistance to clarithromycin in northeast China. World J. Gastroenterol. 2004, 10, 1075–1077.
- Fischbach, L.; Evans, E.L. Meta-analysis: The effect of antibiotic resistance status on the efficacy of triple and quadruple first-line therapies for Helicobacter pylori. Aliment. Pharmacol. Ther. 2007, 26, 343–357.
- Malfertheiner, P.; Megraud, F.; O’Morain, C.A.; Gisbert, J.P.; Kuipers, E.J.; Axon, A.T.; Bazzoli, F.; Gasbarrini, A.; Atherton, J.; Graham, D.Y.; et al. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut 2017, 66, 6–30.
- Kuo, Y.T.; Liou, J.M.; El-Omar, E.M.; Wu, J.Y.; Leow, A.H.R.; Goh, K.L.; Das, R.; Lu, H.; Lin, J.T.; Tu, Y.K.; et al. Primary antibiotic resistance in Helicobacter pylori in the Asia-Pacific region: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2017, 2, 707–715.
- Shrestha, A.B.; Pokharel, P.; Sapkota, U.H.; Shrestha, S.; Mohamed, S.A.; Khanal, S.; Jha, S.K.; Mohanty, A.; Padhi, B.K.; Asija, A.; et al. Drug Resistance Patterns of Commonly Used Antibiotics for the Treatment of Helicobacter pylori Infection among South Asian Countries: A Systematic Review and Meta-Analysis. Trop. Med. Infect. Dis. 2023, 8, 172.
- Bujanda, L.; Nyssen, O.P.; Vaira, D.; Saracino, I.M.; Fiorini, G.; Lerang, F.; Georgopoulos, S.; Tepes, B.; Heluwaert, F.; Gasbarrini, A.; et al. Antibiotic Resistance Prevalence and Trends in Patients Infected with Helicobacter pylori in the Period 2013–2020: Results of the European Registry on H. pylori Management (Hp-EuReg). Antibiotics 2021, 10, 1058.
- Hulten, K.G.; Lamberth, L.B.; Kalfus, I.N.; Graham, D.Y. National and Regional US Antibiotic Resistance to Helicobacter pylori: Lessons From a Clinical Trial. Gastroenterology 2021, 161, 342–344.e1.
- Ho, J.J.C.; Navarro, M.; Sawyer, K.; Elfanagely, Y.; Moss, S.F. Helicobacter pylori Antibiotic Resistance in the United States Between 2011 and 2021: A Systematic Review and Meta-Analysis. Am. J. Gastroenterol. 2022, 117, 1221–1230.
- Jaka, H.; Rhee, J.A.; Ostlundh, L.; Smart, L.; Peck, R.; Mueller, A.; Kasang, C.; Mshana, S.E. The magnitude of antibiotic resistance to Helicobacter pylori in Africa and identified mutations which confer resistance to antibiotics: Systematic review and meta-analysis. BMC Infect. Dis. 2018, 18, 193.
- Megraud, F.; Coenen, S.; Versporten, A.; Kist, M.; Lopez-Brea, M.; Hirschl, A.M.; Andersen, L.P.; Goossens, H.; Glupczynski, Y.; Study Group, p. Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut 2013, 62, 34–42.
- Kwon, Y.M.; Kim, S.J.; Lee, J.G.; Lee, S.P. Effects of prior antibiotic use on clarithromycin resistance in Helicobacter pylori. Helicobacter 2023, 28, e12974.
- Kocsmar, E.; Buzas, G.M.; Szirtes, I.; Kocsmar, I.; Kramer, Z.; Szijarto, A.; Fadgyas-Freyler, P.; Szenas, K.; Rugge, M.; Fassan, M.; et al. Primary and secondary clarithromycin resistance in Helicobacter pylori and mathematical modeling of the role of macrolides. Nat. Commun. 2021, 12, 2255.
- Lim, S.G.; Park, R.W.; Shin, S.J.; Yoon, D.; Kang, J.K.; Hwang, J.C.; Kim, S.S.; Kim, J.H.; Lee, K.M. The relationship between the failure to eradicate Helicobacter pylori and previous antibiotics use. Dig. Liver Dis. 2016, 48, 385–390.
- Shin, W.G.; Lee, S.W.; Baik, G.H.; Huh, K.C.; Lee, S.I.; Chung, J.W.; Jung, W.T.; Park, M.I.; Jung, H.K.; Kim, H.U.; et al. Eradication Rates of Helicobacter pylori in Korea Over the Past 10 years and Correlation of the Amount of Antibiotics Use: Nationwide Survey. Helicobacter 2016, 21, 266–278.
- Munoz-Gomez, P.; Jordan-Castro, J.A.; Abanades-Tercero, M.; Blanco-Gonzalez, J.J.; Andres Esteban, E.M.; Valle-Munoz, J. Macrolide use in the previous years is associated with failure to eradicate Helicobacter pylori with clarithromycin-containing regimens. Helicobacter 2018, 23, e12452.
- Megraud, F. H pylori antibiotic resistance: Prevalence, importance, and advances in testing. Gut 2004, 53, 1374–1384.
- Megraud, F.; Bruyndonckx, R.; Coenen, S.; Wittkop, L.; Huang, T.D.; Hoebeke, M.; Benejat, L.; Lehours, P.; Goossens, H.; Glupczynski, Y.; et al. Helicobacter pylori resistance to antibiotics in Europe in 2018 and its relationship to antibiotic consumption in the community. Gut 2021, 70, 1815–1822.
- Smith, S.; Fowora, M.; Pellicano, R. Infections with Helicobacter pylori and challenges encountered in Africa. World J. Gastroenterol. 2019, 25, 3183–3195.
- Graham, D.Y.; Lee, Y.C.; Wu, M.S. Rational Helicobacter pylori therapy: Evidence-based medicine rather than medicine-based evidence. Clin. Gastroenterol. Hepatol. 2014, 12, 177–186.e3, Discussion e112–173.
- Hwang, J.Y.; Kim, C.; Kwon, Y.H.; Lee, J.E.; Jeon, S.W.; Nam, S.Y.; Seo, A.N.; Han, M.H.; Park, J.H. Dual Clarithromycin and Metronidazole Resistance Is the Main Cause of Failure in Ultimate Helicobacter pylori Eradication. Dig. Dis. 2021, 39, 451–461.
- Graham, D.Y.; Dore, M.P.; Lu, H. Understanding treatment guidelines with bismuth and non-bismuth quadruple Helicobacter pylori eradication therapies. Expert. Rev. Anti-Infect. Ther. 2018, 16, 679–687.
- Graham, D.Y.; Lee, S.Y. How to Effectively Use Bismuth Quadruple Therapy: The Good, the Bad, and the Ugly. Gastroenterol. Clin. N. Am. 2015, 44, 537–563.
- Graham, D.Y.; Megraud, F. Classification system for Helicobacter pylori therapies: Compared and contrasted to traditional infectious disease therapy. Helicobacter 2021, 26, e12773.
- Garcia, M.; Raymond, J.; Garnier, M.; Cremniter, J.; Burucoa, C. Distribution of spontaneous gyrA mutations in 97 fluoroquinolone-resistant Helicobacter pylori isolates collected in France. Antimicrob. Agents Chemother. 2012, 56, 550–551.
- Nishizawa, T.; Suzuki, H. Mechanisms of Helicobacter pylori antibiotic resistance and molecular testing. Front. Mol. Biosci. 2014, 1, 19.
- Miyachi, H.; Miki, I.; Aoyama, N.; Shirasaka, D.; Matsumoto, Y.; Toyoda, M.; Mitani, T.; Morita, Y.; Tamura, T.; Kinoshita, S.; et al. Primary levofloxacin resistance and gyrA/B mutations among Helicobacter pylori in Japan. Helicobacter 2006, 11, 243–249.
- Rimbara, E.; Noguchi, N.; Kawai, T.; Sasatsu, M. Fluoroquinolone resistance in Helicobacter pylori: Role of mutations at position 87 and 91 of GyrA on the level of resistance and identification of a resistance conferring mutation in GyrB. Helicobacter 2012, 17, 36–42.
- Gebeyehu, E.; Bantie, L.; Azage, M. Inappropriate Use of Antibiotics and Its Associated Factors among Urban and Rural Communities of Bahir Dar City Administration, Northwest Ethiopia. PLoS ONE 2015, 10, e0138179.
- Lawson, A.J.; Elviss, N.C.; Owen, R.J. Real-time PCR detection and frequency of 16S rDNA mutations associated with resistance and reduced susceptibility to tetracycline in Helicobacter pylori from England and Wales. J. Antimicrob. Chemother. 2005, 56, 282–286.
- Dadashzadeh, K.; Milani, M.; Rahmati, M.; Akbarzadeh, A. Real-time PCR detection of 16S rRNA novel mutations associated with Helicobacter pylori tetracycline resistance in Iran. Asian Pac. J. Cancer Prev. 2014, 15, 8883–8886.
- Anoushiravani, M.; Falsafi, T.; Niknam, V. Proton motive force-dependent efflux of tetracycline in clinical isolates of Helicobacter pylori. J. Med. Microbiol. 2009, 58, 1309–1313.
This entry is offline, you can click here to edit this entry!
