Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Cytological approaches have long been used in the diagnosis, prognosis, and management of acute myeloid leukemia (AML) and myelodysplastic neoplasms. Technological advances in molecular biology, in particular next-generation sequencing (NGS), have made it possible to establish a molecular list of several gene mutations in AML and MDS, within a matter of days.
- leukemia
- myelodysplastic neoplasms
- next-generation sequencing
1. Introduction
The management of acute myeloid leukemia (AML) and myelodysplastic neoplasia (MDS) has been based, for a long time, on a cytological approach, which was already based on undiscovered molecular bases. We can mention the eosinophilia-associated features of AML4eo, for which an association with inv(16) or an equivalent was often found, without forgetting the clinical and cytological features of acute promyelocytic leukemia (APL) linked to t(15;17) or an equivalent. Among the major technological evolutions, the progress in deoxyribonucleic acid (DNA) sequencing and/or ribonucleic acid (RNA) amplification by a polymerase chain reaction (PCR) has allowed us to refine our classifications of MDS/AML in order to better understand the mechanisms of leukemogenesis, and thus, to be able to better adapt the treatments. However, until now, these tools were more or less used gene-by-gene and target-by-target. The technological evolution of next-generation sequencing (NGS) now allows us, with a delay of a few days compatible with patient management, to obtain a molecular identity card of MDS/AML involving the analysis of mutations in several dozen genes at the same time.
2. Diagnosis
For the European Leukemia Net (ELN), the International Consensus Classification (ICC) and the World Health Organization (WHO) [1,2,3], molecular abnormalities take precedence over the old cytological features defined by the French-American-British (FAB) classification [4], but also over the phenotypic features defined by flow cytometry. The other clinical or biological criteria (treatment-related leukemia, pre-existing myelodysplastic or myelodysplastic/myeloproliferative neoplasms, genetic predisposition) are now only associated with the molecular diagnosis. Although the ICC and WHO classifications are in great part common, some differences can be observed (Table 1). While the ICC still uses the AML-not otherwise specified (NOS) type, the WHO delineates AML by defining genetic abnormalities vs. AML defined by differentiation. Both classifications introduce a section with other defined genetic alterations in order to allow entering novel significant molecular subtypes. Regarding the molecular subtypes, some differences exist, such as a defined section of AML with nucleoporin (NUP)98 rearrangement, RNA-binding motif protein 15 (RBM15), myocardin-related transcription factor-A (MRTFA) fusion, or other fusion genes than Myeloid/Lymphoid or mixed-lineage Leukemia, translocated to 3 (MLLT3) associated to lysine N-methyltransferase 2A (KTM2A) in the ICC classification, which, in contrast, is absent in the WHO classification. The most important change is that the 20% blast cutoff is not required in both classifications to define AML, except for AML with the Breakpoint Cluster Region (BCR): ABL Proto-Oncogene 1, Non-Receptor Tyrosine Kinase (ABL1) in both classifications, for CEBPA mutations in the WHO classification, AML-Myelodysplasia-related (MR) and AML-NOS defined by differentiation. Nonetheless, while the ICC has defined a cut-off at 10% for the AML with defining genetic abnormalities, the WHO has not assigned an arbitrary lower bone marrow blast cut-off if the molecular data correlates with the morphologic findings. Regarding the defining mutations of AML-MR, eight mutations are common to both classifications. The WHO does not retain the Runt-related transcription factor 1 (RUNX1), in contrast to the ICC. Some differences are also present regarding the AML-MR defining cytogenetic abnormalities (11q and 12p deletion in the WHO, not in the ICC, and +8 in the ICC, not retained in the WHO).
Table 1. Comparison of WHO and ICC classifications for AML.
| WHO 2022 | ICI |
|---|---|
| Acute myeloid leukemia with defining genetic abnormalities Acute promyelocytic leukemia with PML::RARA fusion Acute myeloid leukemia with RUNX1::RUNX1T1 fusion Acute myeloid leukemia with CBFB::MYH11 fusion Acute myeloid leukemia with DEK::NUP214 fusion Acute myeloid leukemia with RBM15::MRTFA fusion Acute myeloid leukemia with BCR::ABL1 fusion Acute myeloid leukemia with KMT2A rearrangement Acute myeloid leukemia with MECOM rearrangement Acute myeloid leukemia with NUP98 rearrangement Acute myeloid leukemia with NPM1 rearrangement Acute myeloid leukemia with CEBPA rearrangement Acute myeloid leukemia, myelodysplasia-related Acute myeloid leukemia with other defined genetic alterations |
Acute myeloid leukemia with defining genetic abnormalities Acute promyelocytic leukemia with PML::RARA fusion or other RARA rearrangement Acute myeloid leukemia with RUNX1::RUNX1T1 fusion Acute myeloid leukemia with CBFB::MYH11 fusion Acute myeloid leukemia with DEK::NUP214 fusion XXX Blast phase CML Acute myeloid leukemia with KMT2A rearrangement: MLLT3 Acute myeloid leukemia with MECOM rearrangement XXX Acute myeloid leukemia with NPM1 rearrangement Acute myeloid leukemia with CEBPA rearrangement Acute myeloid leukemia, myelodysplasia-related Acute myeloid leukemia with other defined genetic alterations |
| Acute myeloid leukemia, defined by differentiation Acute myeloid leukemia with minimal differentiation Acute myeloid leukemia without maturation Acute myeloid leukemia with maturation Acute basophilic leukemia Acute myelomonocytic leukemia Acute monocytic leukemia Acute erythroid leukemia Acute megakaryoblastic leukemia |
AML, not otherwise specified XXX XXX XXX XXX XXX XXX XXX |
| MDS-MR defining mutations ASXL1 BCOR EZH2 RUNX1 SF3B1 SRSF2 STAG2 U2AF1 ZRSR2 |
MDS-MR defining mutations ASXL1 BCOR EZH2 XXX SF3B1 SRSF2 STAG2 U2AF1 ZRSR2 |
Since these molecular anomalies do not have the same prognostic values and therapeutic consequences, a hierarchy in the diagnostic classification was proposed in order to allow a clinical use of these data (Figure 1). The most obvious observation is the presence of a t(15;17) translocation, signing APL, that outweighs the other abnormalities in terms of prognosis and treatment, which is in line with the very specific therapeutic approaches (retinoic acid compounds/arsenic trioxide) used in this leukemia subtype.
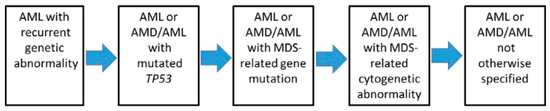
Figure 1. Hierarchical classification of the International Consensus Classification of AML (from [1]).
In first position are the recurrent anomalies (Table 2, + other rearrangements involving the Retinoic Acid Receptor-α (RAR α), KMT2A, or MDS1 and EVI1 complex locus (MECOM)). The presence of these anomalies has another consequence, i.e., the decrease in the diagnostic threshold of AML, which is, in these cases, reduced to only ≥10% of the blasts. Regarding the cytological diagnosis, myeloblasts, monoblasts and megakaryoblasts are considered as blasts, while monoblasts and promonocytes (but not abnormal monocytes) are also classified as blasts in AML subtypes with myelomonocytic or monocytic differentiation. In the setting of APL, promyelocytes are also counted as blasts.
Table 2. Recurrent genetic abnormalities (ELN classification).
| APL with t(15;17)(q24.1;q21.2)/PML::RARA |
| AML with t(8;21)(q22;q22.1)/RUNX1::RUNX1T1 |
| AML with inv(16)(p13.1q22) or t(16;16)(p13.1;q22)/CBFB::MYH11 |
| AML with t(9;11)(p21.3;q23.3)/MLLT3::KMT2A |
| AML with t(6;9)(p22.3;q34.1)/DEK::NUP214 |
| AML with inv(3)(q21.3q26.2) or t(3;3)(q21.3;q26.2)/GATA2, MECOM(EVI1) |
| AML with other rare recurring translocations |
| AML with mutated NPM1 |
| AML with in-frame bZIP mutated CEBPA |
| AML with t(9;22)(q34.1;q11.2)/BCR::ABL1 |
In the second hierarchical position are Tumor Protein (TP)53 mutations (if variant allele frequency [VAF] ≥ 10%), which define a new type of pathology intermediate between acute leukemia and myelodysplastic syndrome (MDS/AML) if blasts are between 10 and 19%, and AML if blasts are ≥20%.
In the third position, the researchers find (with the same distinction AML vs. MDS/AML, according to the percentage of blasts) the mutations related to myelodysplastic syndromes.
In the fourth position, the researchers only find complex karyotypes and/or certain abnormalities related to myelodysplastic syndromes [del(5q)/t(5q)/add(5q),-7/del(7q), +8, del(12p)/t(12p)/add(12p), i(17q),-17/add(17p)/del(17p), del(20q) or idic(X)(q13)]. This downgrading shows the predominant role taken by molecular analysis (by NGS, given the number of anomalies and mutations to be detected) in the classification.
In the fifth position are the leukemias presenting none of the previous characteristics. In addition to the creation of the MDS/AML category, it is important to note the disappearance of the categories of AML with myelodysplasia-related changes (AML-MRC) and treatment-related AML. Indeed, it has been shown that it is the presence of the above-mentioned molecular abnormalities/mutations that impacts the response to treatment, as well as the prognosis, more than these clinical features. This concept is, in fact, not new since it was already known that in AML-MRC or treatment-related categories, karyotypic abnormalities were preponderant in the prognosis.
3. Impact of the Novel Classification on Myelodysplastic Syndrome: The M-IPSS
Among the most interesting impacts of the availability of NGS data is the possibility to refine the prognosis for each patient while evaluating both the classical hematological data (cytopenia, blast counts, age), cytogenetic data and the large mutational analysis data. This led to the elaboration of the Molecular International Prognostic Scoring System (M-IPSS) [30]. The clinical-molecular prognostic model M-IPSS was developed from 2957 MDS patients (604 patients were included in the data set) with a mutation screening of 152 genes. The M-IPSS was secondly tested on a validation cohort of 754 patients. Finally, the score was calculated using hematological parameters, cytogenetic and somatic mutations of 31 genes, leading to six different prognosis categories. In comparison with the previous R-IPSS, 46% of patients were stratified, of whom 74% were upstaged and 26% down-staged. The median OS ranged from 10.6 years (very low M-IPSS) to 1.0 (very high M-IPSS), with death by 4 years, from AML-t, ranging for the same categories from 2.8% to 42.8%, and death without AML from 15.9% to 51.3%. This shows that the M-IPSS not only predicts death by AML but includes other causes, nonetheless, with a less discriminant potency. The TP53multihit mutations, KMT2A (MLL) partial tandem duplication (PTD), and FLT3-ITD correlated with the worse prognosis, while ASXL1, BCOR, EZH2, NRAS, RUNX1, STAG2 and U2AF1 mutations were also associated with adverse risks. The good prognosis of SF3B1 was modified by the presence of co-mutations. Clearly, the analysis of prognosis is not directly feasible by the clinician since it requires taking into account all the mutations and their co-occurrence. Consequently, a useful M-IPSS Web calculator, easily available, has been developed (https://mds-risk-model.com, accessed on 27 October 2022) that also includes missing values and is both applicable for primary and secondary/therapy-related MDS.
4. Measurable Residual Disease (MRD)
The evaluation of MRD can be done with various techniques, mainly using multiparameter flow cytometry (MPFC), real-time polymerase chain reaction (RT-PCR) or NGS [31]. The impact of NGS has to be carefully evaluated. The study by McGowan et al. [31] involved 107 patients and compared 717 MPFC tests to 247 NGS tests. The evaluation of MRD by MPFC was based on the detection of the initial aberrant phenotype by analyzing 500,000 events, while NGS MRD used a panel targeting 141 myeloid-related genes. The patient’s mutations corresponded mainly to FLT3/NPM1, KIT/NRAS/KRAS/PTPN11, GZTA2/CEBPA/WT, TP53/IDH1/IDH2/RUNX1, or DNMT3A/ASXl1/TET, among others. Regarding the 247 instances with both MPFC and NGS MRD tests, there were 197 MPFC+/NGS+ instances, 3 MPFC−/NGS− instances, 44 MPFC−/NGS+ instances and 3 MPFC+/NGS− instances; thus, there was a 19% (47 out of 247) discrepancy between the two methods. Of note, these discrepant results were mainly observed with mutations not associated with adverse outcomes (DNMT3A/ASXL1/TET or others, 17 tests), mutations of unknown significance (14 tests), mutations that were likely pathogenic (3 tests) and 1 patient without mutations. If the researchers exclude these tests, regarding the remaining 131 pairs, 12 are discrepant with 2 MPFC+/NGS− and 10 MPFC−/NGS+. Although the study sample size is limited, these data suggest that NGS may avoid missing low-volume MRD. Nonetheless, the clinical impact of such findings is to be evaluated in controlled trials so that NGS cannot directly impact the treatment that still relies on MPFC and RT-qPCR MRD evaluation. Another impact of NGS is the level of sensitivity of the method used for MRD assessment. The predictive value of NGS vs. MPFC MRD was analyzed and compared in the study by Tsai et al. [32]. This study tested MRD via two different methods in 335 patients with de novo AML in morphological remission (excluding APL) after the first chemotherapy course and after the first consolidation course. The first conclusion was that the study of the common Clonal Hematopoiesis of Indeterminate Potential (CHIP)-related gene mutations (DNMT3A, TET2 and ASXL1) was not informative for MRD assessment. Then, this study showed that the MRD assessment at time 1 by NGS was not predictive of relapse if MPFC MRD was negative but had a poor prognosis value at time 2 even if MPFC was still negative. Due to the rise of specifically targeted therapy, the specific assessment of druggable mutations is of pivotal interest. One of the most common pathogenic mutations detected in AML is FLT3-ITD (25% of patients), with various targeted drugs, such as midostaurin or gilteritinib. The detection of FLT3-ITD MRD is challenging and is mainly done via fragment analysis (FA) with a detection sensitivity of 2%. Recently, the development of novel NGS analysis methods has resulted in a 0.001% sensitivity that was superior to FA analysis [33]. The clinical impact of this higher sensitivity was questioned in the study by Loo et al. [34]. The MRD positivity before the allogeneic transplant was 37% when evaluated by NGS vs. 6.7% by the conventional technique. Of note, the MRD detected by NGS, even at a very low level, had a dismal prognosis, with a 2-year RFS of 78%, 32%, 40% and 0% for NGS MRD level < 0.001 (negative), 0.001–0.1%, 0.1–1% and >1% respectively. In conclusion, FLT3-ITD MRD detection by NGS is an important prognosis factor that can precisely define the relapse risk.
References
1. Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and Management of AML in Adults: 2022 Recommendations from an International Expert Panel on Behalf of the ELN. Blood 2022, 140, 1345–1377, doi:10.1182/blood.2022016867.
2. Arber, D.A.; Orazi, A.; Hasserjian, R.P.; Borowitz, M.J.; Calvo, K.R.; Kvasnicka, H.-M.; Wang, S.A.; Bagg, A.; Barbui, T.; Branford, S.; et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: Integrating Morphologic, Clinical, and Genomic Data. Blood 2022, 140, 1200–1228, doi:10.1182/blood.2022015850.
3. Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th Edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719, doi:10.1038/s41375-022-01613-1.
4. Bennett, J.M.; Catovsky, D.; Daniel, M.T.; Flandrin, G.; Galton, D.A.; Gralnick, H.R.; Sultan, C. Proposed Revised Criteria for the Classification of Acute Myeloid Leukemia. A Report of the French-American-British Cooperative Group. Ann Intern Med 1985, 103, 620–625, doi:10.7326/0003-4819-103-4-620.
30. Bernard, E.; Tuechler, H.; Greenberg, P.L.; Hasserjian, R.P.; Arango Ossa, J.E.; Nannya, Y.; Devlin, S.M.; Creignou, M.; Pinel, P.; Monnier, L.; et al. Molecular International Prognostic Scoring System for Myelodysplastic Syndromes. NEJM Evidence 2022, 1, EVIDoa2200008, doi:10.1056/EVIDoa2200008.
31. F McGowan, P.; D Hyter, S.; Cui, W.; Plummer, R.M.; Godwin, A.K.; Zhang, D. Comparison of Flow Cytometry and Next-Generation Sequencing in Minimal Residual Disease Monitoring of Acute Myeloid Leukemia: One Institute’s Practical Clinical Experience. Int J Lab Hematol 2022, 44, 118–126, doi:10.1111/ijlh.13711.
32. Tsai, C.-H.; Tang, J.-L.; Tien, F.-M.; Kuo, Y.-Y.; Wu, D.-C.; Lin, C.-C.; Tseng, M.-H.; Peng, Y.-L.; Hou, M.-F.; Chuang, Y.-K.; et al. Clinical Implications of Sequential MRD Monitoring by NGS at 2 Time Points after Chemotherapy in Patients with AML. Blood Adv 2021, 5, 2456–2466, doi:10.1182/bloodadvances.2020003738.
33. Lee, J.-M.; Park, S.; Hwang, I.; Kang, D.; Cho, B.S.; Kim, H.-J.; Ahn, A.; Kim, M.; Kim, Y. FLT3-ITD Measurable Residual Disease Monitoring in Acute Myeloid Leukemia Using Next-Generation Sequencing. Cancers (Basel) 2022, 14, 6121, doi:10.3390/cancers14246121.
34. Loo, S.; Dillon, R.; Ivey, A.; Anstee, N.S.; Othman, J.; Tiong, I.S.; Potter, N.; Jovanovic, J.; Runglall, M.; Chong, C.C.; et al. Pretransplant FLT3-ITD MRD Assessed by High-Sensitivity PCR-NGS Determines Posttransplant Clinical Outcome. Blood 2022, 140, 2407–2411, doi:10.1182/blood.2022016567.
This entry is adapted from the peer-reviewed paper 10.3390/cancers15133280
This entry is offline, you can click here to edit this entry!
