Non-canonical, four-stranded nucleic acids secondary structures are present within regulatory regions in the human genome and transcriptome. To date, these quadruplex structures include both DNA and RNA G-quadruplexes, formed in guanine-rich sequences, and i-Motifs, found in cytosine-rich sequences, as their counterparts. Quadruplexes have been extensively associated with cancer, playing an important role in telomere maintenance and control of genetic expression of several oncogenes and tumor suppressors. Therefore, quadruplex structures are considered attractive molecular targets for cancer therapeutics with novel mechanisms of action.
- DNA G-quadruplex
- RNA G-quadruplex
- i-Motif
- cancer
- DNA G4s
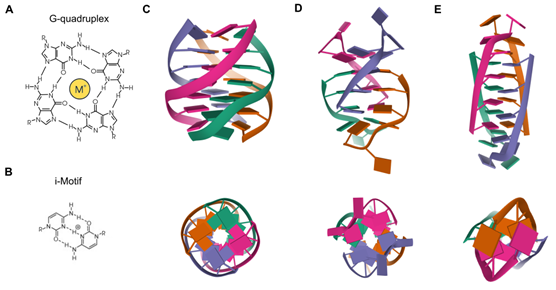
Besides the Watson–Crick double helix, nucleic acids may adopt alternative secondary structures, such as quadruplex structures (Figure 1). DNA G-rich sequences are able to fold into four-stranded secondary structures known as G-quadruplexes (G4), which arose from the self-association of four guanine bases by Hoogsteen hydrogen bonding within a planar G-tetrad. Self-stacking of two or more G-tetrads generates a G4 structure that is further stabilized by monovalent cations, mainly potassium or sodium, and divalent cations including calcium or magnesium [1,2]. G4s are highly polymorphic intra- or intermolecular structures whose topology can be influenced by variations in strand stoichiometry and polarity, as well as by the nature and length of loops and their location in the sequence [3]. G4 stability is affected by numerous factors, including the number of G-tetrads, loop length, topology and the sequence itself, both within the G4 motif and flanking regions [4]. Direct G4-sequencing in purified single-stranded human DNA identified more than 700,000 G4s [5]. However, in situ mapping by G4 ChIP-seq only detected 1000–10,000 endogenous G4s, which accounted for ~1% of those identified by direct G4-seq, possibly owing to chromatin-associated and other proteins that control the formation of these DNA structures [6]. Endogenous G4s have also been visualized in living cells thanks to G4-specific antibody BG4 and were stabilized by a small-molecule ligand such as pyridostatin [7]. In general, G4s are non-randomly distributed through the genome, being mainly clustered in key regulatory sites such as gene promoters, gene bodies and 5′ untranslated regions (5′UTRs) of highly transcribed genes, particularly those related to cancer, amplification of somatic copy number, ribosomal DNA and telomeres [8]. In these genomic locations, G4s are linked to fundamental biological processes such as transcription, replication, genomic instability and telomeres maintenance [9].
Figure 1. Quadruplex structures. (A) Chemical structure of a G4. (B) Chemical structure of an i-Motif. (C) NMR solution structure of a tetrameric parallel G4 (Protein Data Bank: 139D) from different rotation angles. (D) NMR solution structure of an alternating antiparallel tetrameric G4 (Protein Data Bank: 6IMS) from different rotation angles. (E) NMR solution structure of a tetrameric i-Motif (Protein Data Bank: 1YBL) from different rotation angles.
- RNA G4s
While much attention in the G4 field is focused on DNA, these structures are also present in RNA molecules and are known as RNA G-quadruplexes (RNA G4s). In fact, due to the absence of a complementary strand in RNA molecules, it is widely held that G-rich RNA sequences are more prone to the formation of quadruplex structures. However, the thousands of mammalian RNA regions that can fold into G4s in vitro are overwhelmingly unfolded in cells presumably by robust machinery that globally unfolds RNA G4s [10]. RNA G4s rival their DNA counterparts, often displaying enhanced thermodynamic stabilities [11]. Unlike the highly polymorphic DNA G4s structures that depend on the surrounding conditions, RNA G4s were initially believed to adopt a single conformation. The presence of a 2′-hydroxyl group in the ribose sugar results in additional steric constraints and consequently, the topology of RNA G4s is almost exclusively limited to the parallel conformation [12]. Nevertheless, a recent study has opened avenues to consider that RNA G4s can adopt different conformations apart from the parallel one [13]. Compared to DNA G4s, the detection of cellular RNA G4s has been more challenging. RNA G4s were immunodetected using the same DNA G4 recognizing antibody, BG4. This pioneering study provided substantial evidence for the existence of RNA G4s within the cytoplasm of human cells [14]. In the same study, the selectivity and applicability of carboxypyridostatin, as an example of stabilizing ligand targeting RNA G4s within a cellular context, was corroborated [14]. Furthermore, direct RNA G4-sequencing on poly(A)-enriched RNAs mapped G4s structures in more than 3000 human mRNAs. RNA G4s are mostly found in UTRs, but also in coding sequences. Interestingly, G4s are significantly enriched in micro RNAs (miRNAs) and long noncoding RNAs (lncRNAs), as well as in their target sites, suggesting that G4s may influence the interaction of miRNAs and lncRNAs with their target mRNAs [15]. Moreover, human ribosomal RNA was found to form G4 exposed on the ribosomal surface suggesting potential functions on the recruitment of non-ribosomal proteins and/or polysome assembly [16]. Recently, a novel class of small noncoding RNAs induced by stress and named tiRNAs (tRNA-derived Stress-induced RNAs), which play roles in cancer progression, were reported to form G4 and disturb translational initiation [17]. Overall, RNA G4s emerge as pivotal regulators of pre-mRNA processing, miRNAs maturation, RNA turnover, mRNA targeting and translation [18]. Apart from classical DNA G4s and RNA G4s, a new category of intermolecular G4, named DNA:RNA hybrid G-quadruplex (HQ) has been reported to form when two or three G-tracts are present on the non-template DNA strand downstream the transcription start site (TSS). However, HQs formation has only been validated under in vitro conditions so far [19,20].
- i-Motifs
C-rich complementary strands always accompany G-rich sequences in genomic DNA. DNA sequences containing C-stretches have been reported to form intercalated, quadruple-helical structures under acidic conditions, being the resulting structure referred to as i-Motif [21]. The tetrameric structure consists of two parallel duplexes combined in an antiparallel manner through the formation of intercalated hemiprotonated C+:C base pairs. These i-Motifs are inserted in different ways to adopt different topologies known as R-, S- and T-forms. Since i-Motifs are formed by hemiprotonated C+:C base pairs, these structures are more stable in a slightly acidic pH and are able to reversibly fold and unfold just by altering the pH. Interestingly, some i-Motif sequences show stable structures even at neutral pH and ambient temperature [22], being favored under conditions of molecular crowding and negative superhelicity [23]. C-rich regions are common within the human genome and its interest has greatly increased in recent years due to its understanding as functional complementary partners to G4s [24,25]. The generation and characterization of an antibody fragment (iMab) that recognized i-Motif structures with high selectivity and affinity has enabled the detection of i-Motifs in the nuclei of human cells [26]. Such structures are formed in key regulatory regions of the human genome, including promoters and telomeric regions, thus implying their involvement in a variety of replication and transcriptional functions [26].
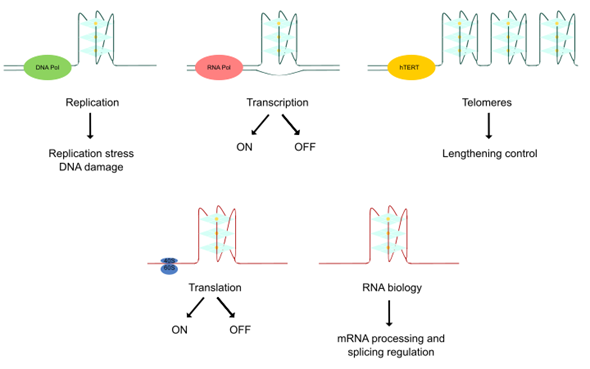
Thanks to the resolution of these non-canonical higher-order structures of nucleic acids and the employment of novel visualization approaches, quadruplex structures have been found to play important roles in several biological events associated with cancer [27]. DNA quadruplexes obstruct the progression of DNA replication forks inducing DNA damage, a hallmark of many tumors. Furthermore, DNA mutations can lead to genomic instability and there is a notable association of quadruplexes with tumoral gene amplification. Lengthening of telomeres is a frequently activated mechanism in cancer and quadruplexes play a key role in telomere biology. Moreover, quadruplexes have been detected in numerous cancer-related genes, interacting with transcription factors or impairing polymerase progression along its template. Thereby, the transcription of different oncogenes and tumor suppressors could be regulated by quadruplex-targeted therapies. In addition, RNA G4s differentially influence translation and mediate recruitment of splicing-associated binding proteins, regulating alternative splicing of numerous important genes in carcinogenesis. Altogether, the cancer-related functions of quadruplexes offer an alternative therapeutic approach in cancer [9,28] (Figure 2).
Figure 2. Biological effects of quadruplex structures. The cancer-related functions of quadruplexes derive from their key roles in replication, transcription, lengthening control of telomeres, translation and RNA biology control.
This review aims to provide an updated report on the literature on quadruplexes regarding cancer therapy. Here we examine the diversity of quadruplexes associated with the six hallmarks of cancer, analyzing their structures and functional implications. Previous reviews have separately covered DNA G4s, RNA G4s and i-Motifs in cancer. Nevertheless, to the best of our knowledge, the present review is the first one gathering together all of these quadruplex structures with relevance in cancer. Since the field of quadruplexes is in continuous change, this review aims to share the current state of knowledge, focusing on the most recent findings in the area.
This entry is adapted from the peer-reviewed paper 10.3390/ijms21238900