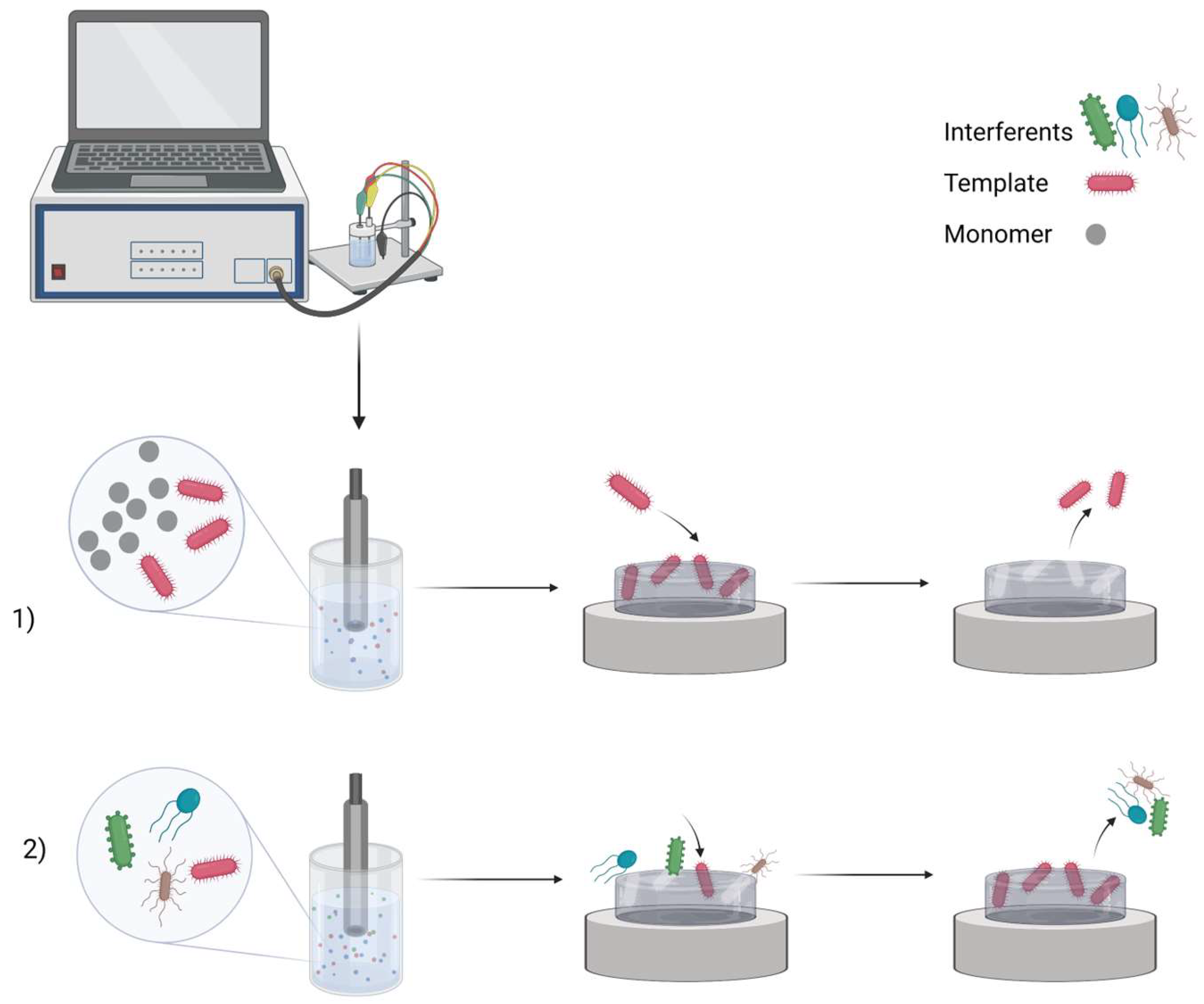
Molecularly imprinted polymer (MIP)-based biosensors have enormous potential for disease detection. Infectious diseases can be detected and identified using MIPs, which are imprinted with whole viruses or specific proteins—biomarkers. Simple detection of the virus can be achieved by whole virus surface imprinting because viruses are easily identified by their morphology and surface properties. Other imprinting techniques and related sensitivity of the prepared MIP-based sensors are bulk imprinting, soft lithography, self-assembly, and the particle core-shell (template immobilization technique). Using MIP-based technology, viruses can be detected by a whole virus, as in the case of the Japanese encephalitis virus imprinted in the tetraethyl orthosilicate or hepatitis A virus imprinted in polydopamine (PDA), virus aptamer (e.g., HIV-1 gene imprinted in poly(o-phenylenediamine on ITO), main protein (e.g., spike protein or NS1 (non-structural protein 1—a specific and sensitive biomarker for dengue virus infection) or HIV-p24 (human immunodeficiency virus p24)), epitope (e.g., glycoprotein 41, gp41 (of related protein to human immunodeficiency virus type 1 (HIV-1))) templates.
- molecularly imprinted polymer (MIP)
- electrochemical sensor
- infectious disease biomarker
- conducting polymer (CP)
- biomarker detection
- COVID-19 detection
- HIV-1 sensor
- Hepatitis C sensor
- sepsis detection
- inflammation detection
- SARS-CoV-2 sensor
- SARS-Cov-2
1. MIP Formation Principles
Figure 1. MIP preparation scheme: (1) imprinting template molecules and then washing them out; (2) rebinding process in the sample. Created with Biorender.com (accessed on 28 May 2023).
2. HIV-1
3. COVID-19
4. Dengue Virus
5. Hepatitis C Virus
6. Nosocomial Infections
| Biomarkers | Polymers and Modifiers | Electrodes | Extraction of Templates | Electrochemical Analysis Methods | LOD, LOQ, LR | Interfering Molecules | Reference |
|---|---|---|---|---|---|---|---|
| HIV-1 | |||||||
| gp41 | PDA | QCM | 5% acetic acid (in H2O) for five times, DI water | X-ray photoelectron spectrometer (XPS) | LOD 2 ng/mL; LR 5–200 ng/mL | [15] | |
| gp120 | Ppy, CNF-Bi, chitosan | GCE | Hyper pure water; methanol and acetic acid solution for 20 min. | CV, DPV | LOD 0.0003 ng/mL; LR 0.002–200 ng/mL | HIV-1 protein p24, human chorionic gonadotropin, carcinoembryonic antigen | [16] |
| COVID-19 | |||||||
| SARS-CoV-2-S spike glycoprotein | Ppy | Pt | Incubation in 0.05 M H2SO4 for 10 min. | Pulsed Amperometric Detection | BSA | [6] | |
| SARS-CoV-2 spike protein | Poly(aminophenylboronic acid) | SPE | 50 mM dithiothreitol for 30 min; 30 min in 10% acetic acid | SWV, CV | LOD 1.12 pg/mL; LR 0–400 fM | SARS-CoV-2 nucleocapsid protein, E2, HSA, IgG | [17] |
| Dengue virus | |||||||
| NS1 | PDA, polysulfone fibres | SPCE | PBS; 500 μg/mL of proteinase K for 2 h in the dark | EIS, CV | LOD 0.3 ng/mL; LR 1–200 ng/mL | FBS, lysozyme | [19] |
| NS1 | Poly(G03TCOOH), gold | QCM | Potential washing (−0.7 V) 0.1 M tetrabutylammonium hexafluorophosphate in acetonitrile | EIS | LOD 0.056 μg/mL; LR 0.2 to 10 μg/mL | angiotensin II human, glycyl glycine, bovine serum albumin, fibrinogen | [20] |
| Hepatitis C virus | |||||||
| HCV surface protein E2 | PmPD | SPE | PBS with 50 mM dithiothreitol for 30 min, 10% acetic acid solution on vortex for 30 min | DPV | LOD 0.46 pg/mL; LR 0.01–50 ng/mL; LOQ 15.3 × 10−5 ng/mL | HSA, IgG, CD81 | [21] |
| HCV core antigen | PDA, MWCNTs- Chit nanocomposite | GCE | Water, overnight in 5% v/v acetic acid and 1% w/v cetyl trimethyl ammonium bromide in water with stirring | CV, DPV, EIS | LOD 1.67 fg/mL; LR 5.0 fg/mL to 1.0 pg/mL; | [22] | |
| Nosocomial infections | |||||||
| K. pneumoniae | Ppy | ITO | DI water, ethanol | CV, DPV | LOD 1.352 CFU/mL; LR 1–105 CFU/mL | uric acid, K+, Mg++, urea, Lactobacillus, E. coli | [23] |
| K. pneumoniae | Poly(MAM:AAM:NVP), graphene oxide | AuSPE | 10% acetic acid for 30 min, water at 50 °C for 30 min | CV | LOD 0.012 CFU/mL; LOQ 1.61 CFU/mL; LR 101–105 CFU/mL | E. faecalis, P. aeruginosa | [13] |
| P. aeruginosa | PDA, AuNPs | GCE | Solution containing SDS 0.01 M and 5% HNO3 in water | CV, EIS, DPV | LOD 1 CFU/mL; LR 10–107 CFU/mL | Shigella flexneri, Salmonella enteritidis, E. coli, K. pneumonia | [24] |
| L. monocytogenes | Ppy | SPCE | 10% acetic acid, or sulfuric acid, or L-lysin, or trypsin | PAD | LOD 70 CFU/mL, LOQ 210 CFU/mL, LR 300–6700 CFU/mL. | [26] | |
This entry is adapted from the peer-reviewed paper 10.3390/bios13060620
References
- Ratautaite, V.; Topkaya, S.N.; Mikoliunaite, L.; Ozsoz, M.; Oztekin, Y.; Ramanaviciene, A.; Ramanavicius, A.; Molecularly imprinted polypyrrole for DNA determination. Electroanalysis 2013, 25, 1169–1177, .
- Baleviciute, I.; Ratautaite, V.; Ramanaviciene, A.; Balevicius, Z.; Broeders, J.; Croux, D.; McDonald, M.; Vahidpour, F.; Thoelen, R.; Ceuninck, W.D.; et al. Evaluation of theophylline imprinted polypyrrole film. Synth. Met. 2015, 209, 206–211, .
- Ratautaite, V.; Janssens, S.D.; Haenen, K.; Nesládek, M.; Ramanaviciene, A.; Baleviciute, I.; Ramanavicius, A.; Molecularly imprinted polypyrrole based impedimetric sensor for theophylline determination. Electrochim. Acta 2014, 130, 361–367, .
- Ratautaite, V.; Nesladek, M.; Ramanaviciene, A.; Baleviciute, I.; Ramanavicius, A.; Evaluation of histamine imprinted polypyrrole deposited on boron doped nanocrystalline diamond.. Electroanalysis 2014, 26, 2458–2464, .
- Ratautaite, V.; Boguzaite, R.; Brazys, E.; Ramanaviciene, A.; Ciplys, E.; Juozapaitis, M.; Slibinskas, R.; Bechelany, M.; Ramanavicius, A.; Molecularly imprinted polypyrrole based sensor for the detection of SARS-CoV-2 spike glycoprotein.. Electrochim. Acta 2022, 403, 139581, .
- Ratautaite, V.; Plausinaitis, D.; Baleviciute, I.; Mikoliunaite, L.; Ramanaviciene, A.; Ramanavicius, A.; Characterization of caffeine-imprinted polypyrrole by a quartz crystal microbalance and electrochemical impedance spectroscopy. Sens. Actuator B-Chem. 2015, 212, 63-71, .
- Plausinaitis, D.; Ratautaite, V.; Mikoliunaite, L.; Sinkevicius, L.; Ramanaviciene, A.; Ramanavicius, A.; Quartz crystal microbalance-based evaluation of the electrochemical formation of an aggregated polypyrrole particle-based layer. Langmuir 2015, 31, 3186-3193, .
- Plausinaitis, D.; Sinkevicius, L.; Samukaite-Bubniene, U.; Ratautaite, V.; Ramanavicius, A.; Evaluation of electrochemical quartz crystal microbalance based sensor modified by uric acid-imprinted polypyrrole.. Talanta 2020, 220, 121414, .
- Balciunas, D.; Plausinaitis, D.; Ratautaite, V.; Ramanaviciene, A.; Ramanavicius, A.; owards electrochemical surface plasmon resonance sensor based on the molecularly imprinted polypyrrole for glyphosate sensing. Talanta 2022, 241, 123252, .
- Ratautaite, V.; Brazys, E.; Ramanaviciene, A.; Ramanavicius, A.; Electrochemical sensors based on L-tryptophan molecularly imprinted polypyrrole and polyaniline. J. Electroanal. Chem. 2022, 917, 116389, .
- Ratautaite, V.; Samukaite-Bubniene, U.; Plausinaitis, D.; Boguzaite, R.; Balciunas, D.; Ramanaviciene, A.; Neunert, G.; Ramanavicius, A.; Molecular imprinting technology for determination of uric acid. Int. J. Mol. Sci. 2021, 22, 5032, .
- Ramanaviciene, A.; Ramanavicius, A.; Molecularly imprinted polypyrrole-based synthetic receptor for direct detection of bovine leukemia virus glycoproteins. Biosens. Bioelectron. 2004, 20, 1076-1082, .
- Pintavirooj, C.; Vongmanee, N.; Sukjee, W.; Sangma, C.; Visitsattapongse, S.; Biosensors for Klebsiella pneumoniae with molecularly imprinted polymer (MIP) technique.. Sensors 2022, 22, 4638, .
- Ramanavicius, S.; Jagminas, A.; Ramanavicius, A.; Advances in molecularly imprinted polymers based affinity sensors (review). Polymers 2021, 13, 974, .
- Lu, C.-H.; Zhang, Y.; Tang, S.-F.; Fang, Z.-B.; Yang, H.-H.; Chen, X.; Chen, G.-N.; Sensing HIV related protein using epitope imprinted hydrophilic polymer coated quartz crystal microbalance. Biosens. Bioelectron. 2012, 31, 439–444, .
- Ma, Y.; Liu, C.; Wang, M.; Wang, L.-S.; Sensitive electrochemical detection of gp120 based on the combination of NBD-556 and gp120. Talanta 2019, 196, 486–492, .
- Ayankojo, A.G.; Boroznjak, R.; Reut, J.; Öpik, A.; Syritski, V.; Molecularly imprinted polymer based electrochemical sensor for quantitative detection of SARS-CoV-2 spike protein. Sens. Actuator B-Chem. 2022, 353, 131160, .
- Liustrovaite, V.; Drobysh, M.; Rucinskiene, A.; Baradoke, A.; Ramanaviciene, A.; Plikusiene, I.; Samukaite-Bubniene, U.; Viter, R.; Chen, C.-F.; Ramanavicius, A.; et al. Towards an electrochemical immunosensor for the detection of antibodies against SARS-CoV-2 spike protein. J. Electrochem. Soc. 2022, 169, 037523, .
- Arshad, R.; Rhouati, A.; Hayat, A.; Nawaz, M.H.; Yameen, M.A.; Mujahid, A.; Latif, U.; MIP-based impedimetric sensor for detecting Dengue fever biomarker. Appl. Biochem. Biotechnol. 2020, 191, 1384-1394, .
- Buensuceso, C.E.; Tiu, B.D.B.; Lee, L.P.; Sabido, P.M.G.; Nuesca, G.M.; Caldona, E.B.; del Mundo, F.R.; Advincula, R.C.; Electropolymerized-molecularly imprinted polymers (E-MIPS) as sensing elements for the detection of dengue infection. Anal. Bioanal. Chem. 2022, 414, 1347-1357, .
- Antipchik, M.; Reut, J.; Ayankojo, A.G.; Öpik, A.; Syritski, V.; MIP-based electrochemical sensor for direct detection of hepatitis C virus via E2 envelope protein. Talanta 2022, 250, 123737, .
- Ghanbari, K.; Roushani, M.; A nanohybrid probe based on double recognition of an aptamer MIP grafted onto a MWCNTs-Chit nanocomposite for sensing hepatitis C virus core antigen. Sens. Actuator B-Chem. 2018, 258, 1066-1071, .
- Sharma, R.; Lakshmi, G.B.V.S.; Kumar, A.; Solanki, P.; Polypyrrole based molecularly imprinted polymer platform for Klebsiella pneumonia detection. ECS Sens. Plus 2022, 1, 010603, .
- Sarabaegi, M.; Roushani, M.; Rapid and sensitive determination of Pseudomonas aeruginosa by using a glassy carbon electrode modified with gold nanoparticles and aptamer-imprinted polydopamine. Microchem. J. 2021, 168, 106388, .
- Tokonami, S.; Nakadoi, Y.; Takahashi, M.; Ikemizu, M.; Kadoma, T.; Saimatsu, K.; Dung, L.Q.; Shiigi, H.; Nagaoka, T.; Label-free and selective bacteria detection using a film with transferred bacterial configuration. Anal. Chem. 2013, 85, 4925–4929, .
- Liustrovaite, V.; Pogorielov, M.; Boguzaite, R.; Ratautaite, V.; Ramanaviciene, A.; Pilvenyte, G.; Holubnycha, V.; Korniienko, V.; Diedkova, K.; Viter, R.; et al. Towards electrochemical sensor based on molecularly imprinted polypyrrole for the detection of bacteria-Listeria monocytogenes. Polymers 2023, 15, 1597, .