Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biochemistry & Molecular Biology
Guanine nucleotide-binding protein-like 3 (GNL3) and proliferation-associated protein 2G4 (PA2G4) are molecules involved during metaphase-to-anaphase transition and growth regulation. GNL3 and PA2G4 have been found to be overexpressed in several human cancers, including prostate cancer. Clinical data suggest that GNL3 and PA2G4 could be developed as prognostic biomarkers of clinical significance in prostate cancer.
- prostate cancer
- biomarkers
- overall survival
- TCGA database
- prognostics
1. Introduction
Prostate cancer is a prevalent disease in males worldwide with an existing rate of 29% for all diagnosed cancers and the fifth leading cause of cancer-related deaths [1]. According to Globocan 2020, approximately 1.14 million prostate cancer cases were diagnosed and 0.375 million deaths occurred in 2020 [2]. Prostate cancer is a heterogeneous disease, ranging from remarkably low-aggressive, organ-confined to high-aggressive, non-organ confined lethal phenotypes. The therapeutic decision and survival outcome of prostate cancer is dependent on appropriate patient stratification to different risk groups; therefore, it is very important to differentiate between indolent and aggressive diseases. Clinical diagnosis and prognosis of prostate cancer is currently based on digital rectal examination (DRE), serum levels of prostate-specific antigen (PSA), and pathologic Gleason score. The PSA test as a screening tool for prostate cancer was first approved by the Food and Drug Administration in 1986, and is still controversial because of high false-positive rates and the risks associated with biopsies and over-treatment [3]. As such, PSA is a non-specific biomarker for prostate cancer, and its expression has also been reported in other organs such as the adrenals, small intestine, kidney, and salivary tissue [4]. There is a strong debate regarding PSA as a diagnostic and prognostic marker since it is unable to differentiate between indolent and aggressive forms of prostate cancer. This is evidenced by the fact that many men harbor aggressive prostate cancer while displaying low levels of serum PSA. Moreover, the Gleason grading system used with prostate biopsy specimens to evaluate the clinical progression of men with prostate cancer needs further refinement for more accurate grade stratification. Together with these parameters, it is especially important to focus on other types of molecular markers that can support clinical outcomes and disease prognostication [5,6].
2. Biological Pathways and Protein–Protein Interaction Analysis
Previous analysis identified GNL3 and PA2G4 as the lead genes after Kaplan–Meier survival and ROC analysis. Further, these two gene sets were chosen to study their involvement in biological pathways, protein–protein interaction (PPI), and co-expression pattern. Researchers utilized FunRich, an offline software to determine the involvement of GNL3 and PA2G4 in different biological signaling pathways by applying a statistically significant p value less than 0.05. PPI and co-expression analysis was performed using PPI string database. Researchers observed that GNL3 and PA2G4 were enriched in the regulation and co-regulation of androgen receptor (AR) activity, androgen receptor-mediated signaling, regulation of β-catenin signaling, and Wnt signaling pathways (Figure 1A–C).
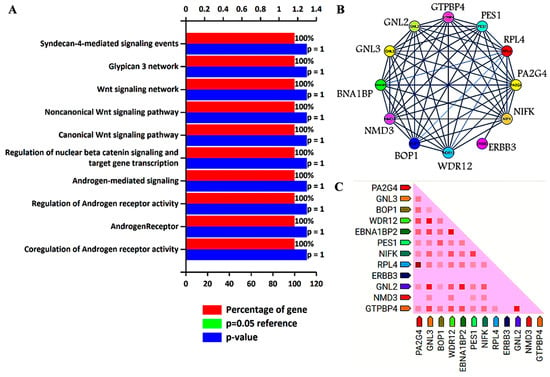
Figure 1. Biological signaling pathway enrichment and protein–protein interaction analysis of GNL3 and PA2G4 genes. (A) Top ten biological signaling pathways of GNL3 and PA2G4. Green bar is invisible as it overlaps with the blue bar. (B) Protein–protein interaction of GNL3, and PA2G4. GNL3 and PA2G4 are displayed in yellow circle. (C) Observed co-expression of GNL3 and PA2G4 in humans.
The top ten biological pathways of GNL3 and PA2G4 with their gene count percentage and significant –log10 p value include syndecan-4 mediated signaling, glypican network, canonical and non-canonical Wnt signaling, β-catenin signaling, and androgen receptor (AR) signaling pathways (Figure 1A). Accumulating data suggests that AR-mediated signaling pathways play a crucial role in the development, progression, and resistance to antiandrogen therapy of prostate cancer. Accumulating evidence indicates a significant role of androgen receptor splice variants in mediating resistance of castration-resistant prostate cancer to current therapies and in predicting therapeutic responses [36,37]. Similarly, Wnt signaling components also play a significant role in prostate tumorigenesis and promote resistance against androgen deprivation therapy [38,39]. Both canonical and non-canonical Wnt signaling pathways regulate several developmental and biological processes including cell proliferation, self-renewal, and stem cell differentiation. In the non-canonical Wnt signaling pathway, Wnt5a, Wnt5b, and Wnt11 ligands bind to a panel of diverse receptors to activate Wnt signaling, including receptors of the Frizzled family and other mediators such as tyrosine-protein kinase transmembrane receptor (ROR1, ROR2 or RYK) [40]. Binding of these non-canonical Wnt ligands can activate multiple intracellular pathways including the planar cell polarity and calcium signaling pathways. The non-canonical Wnt signaling pathway plays a role in prostate cancer progression to an AR-indifferent or neuroendocrine phenotype where the Wnt secretion mediator, Wntless is recognized as a major driver of neuroendocrine-differentiated prostate cancer characterized by aggressive tumor growth [41]. These Wnt prostate tumors express minimal to low levels of AR and reduced PSA. The canonical Wnt signaling is dependent on β-catenin as an effector of Wnt proteins, and its high level induces tumorigenesis. In the absence of extracellular Wnt signals, cytoplasmic β-catenin is phosphorylated by glycogen synthase kinase 3 (GSK3) as part of a destruction complex including adenomatous polyposis coli (APC) and axin proteins. The phosphorylated β-catenin is then ubiquitinated and degraded. Wnt signaling inhibits this process leading to the accumulation of β-catenin in the nucleus by enabling the formation of transcriptionally active complexes [42]. Interaction of β-catenin and its crosstalk with AR has been well documented in prostate cancer. AR binds β-catenin directly to stimulate AR-mediated gene transcription that provides a growth advantage engaging downstream targets such as c-Myc and cyclin D1, even at the castration levels of androgens [43]. The PPI network analysis further revealed that GNL3 and PA2G4 have greater interaction with their co-expressed/co-occurred proteins (Figure 1B,C). In the co-expression analysis, researchers found that GNL3 protein strongly co-expressed with WDR12 and GNL2 proteins. Similarly, PA2G4 protein expression was closely associated with the expression of RPL4 protein (Figure 1C).
The coiled-coil-helix-coiled-coil-helix domain containing 8 (CHCHD8) is a putative COX assembly factor known as cytochrome c oxidase assembly factor 4 homolog (CoA4). COA4 is a newly identified CcO assembly factor which is a twin CX(9)C motif mitochondrial protein localized in the intermembrane space linked with the inner membrane of mitochondria. Its transport into intermembrane space depends on the MIA40 trans-site receptor machinery [44,45]. It is well-known that the mitochondria organelles are a leading source of cellular energy and reactive oxygen species (ROS) [45]. Preclinical and clinical studies have demonstrated that increased levels of ROS, especially free radicals, cause oxidative damage in DNA, proteins, and lipids which lead to the pathogenesis and the progression of prostate cancer. Cytochrome c oxidase (CcO) is an enzyme in the mitochondrial respiratory chain that powers cellular energy production as ATP. CcO enzyme is a tightly regulated protein that is involved in mitochondrial mediated oxidative metabolism, phosphorylation, and ATP formation. In general, the CoA4 protein plays a role in CcO assembly for the mitochondrial respiratory chain. Therefore, deletion and a mutation in the CoA4 protein impair the assembly of CcO, which induces the amplification of hydrogen peroxide production and prevents cell proliferation in a normal condition [46]. Moreover, it has also been shown that CoA4-lacking cells suppress CcO activity [25]. The suppressed activity of CcO has been implicated in the metabolic shift towards glycolysis, and defects in the assembly of the CcO complex lead to the induction of Ca2+/Calcineurin-mediated retrograde signaling. Ca2+/Calcineurin-mediated retrograde signaling can activate PI3-kinase, IGF1R, and AKT. It is well documented that these proteins are involved in the oncogenic transformation and cancer progression [47]. However, the role of CoA4 is limited in cancer. Krobthong et al. (2022) reported that cancer-promoting proteins, including CoA4, were downregulated in the natural peptides treated A549 lung cancer cells compared to the non-treated cells. The authors describe that natural peptides have anti-proliferative and anti-metastatic activities by suppressing cancer-promoting proteins and further reported that the anti-oxidative activity of natural peptides may be attributed to higher expression of ROS-reducing proteins [48]. This study indicates that CoA4 may be involved in ROS production.
The Guanine nucleotide-binding protein-like 3 (GNL3) is alternatively known as nucleostemin. In general, GNL3 stabilizes the telomeric repeat binding factor 1 (TRF1) protein during the processes of mitosis and stimulates the metaphase-to-anaphase transition. GNL3-mediated stabilization of TRF1 controls the telomere and cell cycle progression [49]. Telomeres are the protective structures of chromosome ends that are gradually shortened by each cell division, eventually leading to cellular senescence. Malignant cells maintain the telomere length for unlimited growth by telomerase reactivation or a recombination-based mechanism. Therefore, telomere length has emerged as an emerging therapeutic target in majority of human cancers. Elevated expression of GNL3 in various cancers has been detected. Several studies reported that elevated expression of GNL3 protein promotes cell proliferation, invasion, migration, and epithelial-to-mesenchymal transition in several cancers, including prostate cancer [50,51,52]. It has been reported that loss of GNL3 expression inhibits cell proliferation, migration, invasion and induces apoptosis in various types of cancer cells [53,54]. Recently, Zhang et al. (2022) found the overexpression of GNL3 protein promotes malignant behavior of liver cancer cells. The authors further demonstrated that knockdown of GNL3 inhibits proliferation, migration, and invasion of liver carcinoma cells. These results highlight that aberrant expression of GNL3 is associated with poor overall survival of hepatocellular carcinoma patients [44]. Another in vitro study found that overexpression of GNL3 protein accelerates epithelial-to-mesenchymal transition and decreased expression of GNL3, thereby lowering growth, migration, and invasion of osteosarcoma cells [54]. Similarly, Dai et al. (2021) showed that upregulated expression of GNL3 promotes non-Hodgkin lymphoma progression by stimulating the oncogenic Wnt/β-catenin signaling [55]. Another study found that elevated expression of GNL3 with STAT3 activation is an early process in the progression of low-grade dysplasia squamous cell carcinoma [56]. Sami et al. (2019) demonstrated that nucleostemin (GNL3) protein is a predicted biomarker in the most aggressive phenotype of breast cancer [57]. Lin et al. (2019) revealed that higher nucleostemin expression is associated with poor progression-free survival in triple-negative/basal-like breast cancers [58]. Few other studies found upregulation of GNL3 in prostate cancer. Liu et al. (2009) reported that nucleostemin acts as a critical G1/S barrier regulator, and its higher expression promotes prostate cancer progression [59]. Another study reported that higher expression of nucleostemin at the mRNA and protein level in prostate cancer cells and tissues and that decrease in its expression inhibits PC-3 cell proliferation [60]. Furthermore, a study reported that higher nucleostemin expression enhances prostate cancer tissues’ malignant behavior [35]. Various in vitro studies reported that decreased expression of GNL3 noticeably reduces proliferation in prostate cancer cells [52,60,61,62].
The proliferation-associated protein 2G4 (PA2G4) is an ErbB3-binding protein 1 (EBP1). EBP1 (PA2G4) is ubiquitously expressed in the nucleus and cytoplasm of malignant and non-malignant cells [63,64]. Cytoplasmic Ebp1 protein binds with the cytoplasmic domain of ErbB3 protein and leads to activation of ErbB3 that has been linked with several human cancers, and it has also been reported that ErbB3 signaling plays a crucial role in prostate cancer [63,65]. However, elevated expression of the PA2G4 gene has also been associated with the prognosis of various cancers including hepatocellular carcinoma, nasopharyngeal carcinoma, neuroblastoma, breast, and pancreatic cancers [63,66]. Several studies reported that higher expression of PA2G4 protein promotes cell proliferation, invasion, and migration and inhibits apoptosis in various cancer cells. Recently, Sun et al. (2022) found differential expression of PA2G4 in hepatocellular carcinoma compared to normal liver tissue and reported that the overexpression of PA2G4 accelerates epithelial-to-mesenchymal transition in hepatocellular carcinoma via stabilizing the mRNA of FYN oncogene and its overexpression is associated with poor prognosis of hepatocellular carcinoma patients [67]. Another study reported that higher expression of PA2G4 protein is responsible for poor outcomes in nasopharyngeal carcinoma. Further studies demonstrated that elevated expression of PA2G4 is closely associated with poor survival of nasopharyngeal carcinoma patients and acts as an independent prognostic indicator in the survival of these patients [68]. Liu et al. (2015) reported that high levels of Ebp1 are positively associated with the TNM stages of cervical cancer and involved in lymphatic metastasis [69]. Hou et al. (2021) reported that silencing of the PA2G4 gene repressed cell viability and induced apoptosis by activating the expression of caspase-3 and caspase-9 in glioblastoma cells [70]. Few studies reported overexpression of Ebp1 (PA2G4) protein in prostate cancer. It is known that the ErbB3 protein receptor is a crucial regulator in prostate cancer progression, and it has been demonstrated that binding of Ebp1 with ErbB3 protein is involved in their activation. Loss of expression of Ebp1 protein leads to deactivation of ErbB2/3 signaling, which significantly suppresses the growth of castration-resistant prostate cancer xenografts [71,72,73]. Ectopic expression of Ebp1 regulates cellular proliferation and enhances differentiation in prostate cancer cells [74]. Gannon et al. (2008) demonstrated that high expression of Ebp1 is directly associated with prostate cancer and progressively involved in the transition from the hormone-sensitive to the hormone-refractory stage. Furthermore, in vitro and in vivo study revealed that overexpression of Ebp1 causes therapeutic resistance, and its downregulation increases the sensitivity towards lapatinib in prostate cancer [75].
The Ribosomal RNA processing9 (RRP9) is a U3 small nucleolar RNA binding protein. The Rrp9 protein consists of a WD repeat domain and an N-terminal region, and these WD repeat domains are commonly involved in the interactions between the proteins [76]. Besides WD repeat domain, RRP9 also has another domain that is β-propeller that is composed of seven WD subdomains and mediate the interaction of RRP9 within the small subunit (SSU)-processome. The SSU-processome is a large ribonucleoprotein that is required for the assemblage of the SSU of the ribosomes and for the activation of the 18S rRNA, and the SSU processome is basically responsible for cell survival. RRP9 protein is involved in the functioning of U3 small nucleolar RNA, and U3 and RRP9 are responsible for 18S rRNA production by the SSU processome complex required for early pre-rRNA cleavages at the sites of A0, A1, and A2 [77]. Mutated RRP9 suppresses cell growth, while WT RRP9 restored yeast cell growth [77]. A recent study reported that a RRP9 neddylation deficit prevents pre-rRNA processing and leads to the downregulation of ribosomal biogenesis. Some studies suggested that overexpression of RRP9 enhances tumor cell proliferation, colony formation, and cell migration [78]. Moreover, ribosomal RNA biogenesis has been associated with various human malignancies [79]. Zhang et al. (2022) explored the role of U3 small nucleolar RNA binding protein RRP9 in pancreatic cancer. They reported that the RRP9 protein activates the AKT signaling pathway by binding with the DNA binding area of IGF2BP1 in pancreatic cancer cells, accelerating cancer progression, inhibiting apoptosis, and causes resistance against gemcitabine via decrease in DNA damage. It is reported that the expression of RRP9 is inversely correlated with the prognosis of pancreatic cancer patients [79]. Du et al. (2022) explored the oncogenic role of RRP9 in colon cancer. They found that elevated expression of RRP9 is involved in colorectal cancer tumorigenesis and its progression, and that the loss of RRP9 inhibits cell proliferation and migration, promotes tumor cell senescence, and halts tumor growth in nude mice xenografts [78].
This entry is adapted from the peer-reviewed paper 10.3390/cancers15102723
This entry is offline, you can click here to edit this entry!
