Thanks so much for your check. We sincerely hope you may create this entry, since you are the expert in this academic research. You can click the “submit” button to upload it. We will help you layout after you submit it. Moreover, we will link your article at the entry, and more scholars and students can look through it.
1. Introduction
Research in the past decade has revealed the lofty role of alterations in replicative DNA polymerases (pols) in sporadic and hereditary cancer [
1,
2]. The predisposition to tumorigenesis is attributed to the low fidelity of DNA replication by inaccurate pol versions [
3,
4]. Among the replicative B-family enzymes, pol ε stands out. The alterations in the proofreading exonuclease domain caused by mutations in the
POLE gene (see for the nomenclature of DNA polymerase subunits in humans and in yeast and mouse models) are proven to be causative factors in the etiology of the malignant transformation (), with predominant, but not exclusive prevalence, in colon and endometrial cancers. The review analyzes how the modern understanding of the replication fork based on the synthesis of information gained in model systems and genomics of tumors may explain the peculiarities of the connection of pols and cancer in humans.
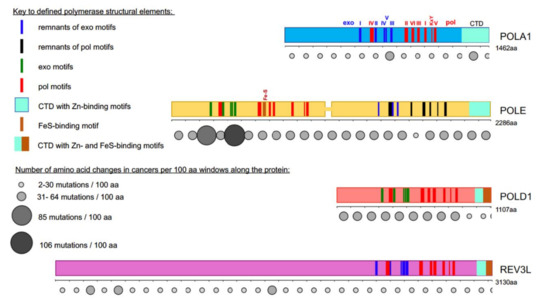
Figure 1. Most cancer-associated mutations affect the catalytically active half of POLE. Colored bars represent the main subunits of DNA pols, a catalytic subunit of pol α, POLA1, in light blue; of pol ε, POLE, in yellow; of pol δ, POLD1, in red; and pol ζ, REV3L, in purple. Note that POLE is a tandem of active pol (
N-terminal half) and inactive pol (C-terminal half) [
5,
6]. Evolutionarily conserved motifs characteristic for all exonuclease (exo) domains, are labeled I-V in green and pol domains are labeled I-VI and KxY in red [
5,
6,
7,
8,
9,
10,
11,
12]. The order of the motifs along all four proteins is the same, but they occupy different parts of the whole protein. For example, REV3L has a very long
N-terminal part not related to pols. In POL1, REV3L, and the C-terminal half of POLE, the exonuclease motifs are inactivated during evolution; they are shown in blue. Inactivated pol motifs in the C-terminal half of POLE are shown in black. The key for these and other elements of the pol primary structure is in the left upper quarter of the figure. Rows of circles of different sizes and shades of grey below the catalytic pol subunits represent the number of missense mutations found in tumors along the protein regions in 100 amino acid increments. Variants were collected from the cBioPortal database from a curated non-overlapping collection of tumor genomes (
https://www.cbioportal.org/ (
cbioportal.org)). A guide explaining the relation between size and intensity of grey to the number of mutations found in the database in the 100 amino acids interval is on the left lower quarter of the figure.
2. Progress on the Structure-Function of B-Family DNA Polymerases and Organization of the Replication Fork
The past ten years brought groundbreaking discoveries about B-family DNA pols. Currently, with the help of X-ray crystallography and the improvement of cryo-EM resolution, we understand the atomic details of the structures of catalytic cores and whole complexes () of yeast and human () primase-pol α [
56,
57,
58], yeast and human pol δ [
59,
60,
61], yeast pol ε alone or in complex with CMG [
62,
63,
64,
65], yeast pol ζ [
66] ().
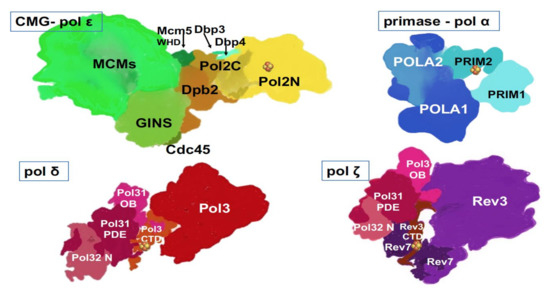
Figure 3. Multi-subunit replicative DNA pols. Artistic representations were made based on crystal and cryo-EM structures and models of human primase-pol α [
58], yeast pol ε [
65,
67], yeast pol δ [
60], yeast pol ζ [
66]. Two latter structures were determined with a truncated third subunit (), Pol 32, without the C-terminal part, and thus, this part is missing from our drawings. Fe-S cluster (
) is present in each of the four pol complexes.
As we can see from the list, two human pols’ structures can only be modeled based on solved yeast counterparts, thus making the solution of structures of human pols a high priority. The structures of yeast and human pols appear to be similar in general features but differ in nuances. For example, human pol δ has an additional small subunit, p12 () hypothesized to regulate pol δ activity during normal replication versus conditions of DNA damage or replicative stress [
61,
68]. The catalytic subunit of human pol ζ has extended the
N-terminal part of unknown significance (, and ). Structural and functional studies helped understand transactions in the active site of polymerases and within the pol complexes. Examples of success are the basis of RNA primer synthesis by primase and transfer of RNA primer 3′-end into pol α active site to start DNA synthesis; the reasons for the high fidelity of pols δ and ε; and the ability of pol ζ to extend mismatches or unpaired DNA ends found opposite lesions.
One exciting finding is that all DNA pols coordinate Fe-S clusters, known regulatory/structural elements of various proteins [
69], alluding to the connection of iron metabolism in mitochondria to replication and novel opportunities for regulation of pol reactions ( and ) [
70,
71]. The cluster can accept or donate electrons and might be involved in sensing the redox potential of cells and DNA damage [
72,
73]. The first finding was the detection of the Fe-S cluster in the second subunit of archaeal and yeast primase [
74], which was proven to play a seminal role in the primer synthesis by human enzyme [
58,
75,
76,
77]. Then, Fe-S clusters were found and verified in C-terminal regions of yeast and human pols δ and ζ ( and
) and were shown to be necessary for pol function [
70,
78,
79]. The Fe-S cluster in the catalytic subunit of pol ε was found in an unusual location: in the
N-terminal half in the vicinity of pol II motif (), structurally characterized, and shown to be necessary for pol but not exo activity in functional assays [
80,
81]. A recent study revealed the unique sensitivity of pol ε to suppression of Fe-S biosynthesis in basal-like breast cancer cell lines [
55].
Another sensational discovery was the sharing of subunits between pols δ and ζ [
78], , . For quite an extended period, pol ζ was referred to as a two subunit enzyme consisting of catalytic Rev3 and accessory subunit Rev7 [
82], later found to be one REV3 to two REV7 subunit complex [
66,
83]. The two-subunit complex possessed quite low and variable activity [
84,
85,
86]. The pol δ’s two accessory subunits appeared to be two additional subunits of pol ζ necessary for the full activity [
78,
87,
88,
89]. It appeared that the inconsistent activity of former “two-subunit” preps resulted from uncontrolled traces of a genuine four subunit enzyme [
88]. The role of such subunit sharing between the main replicative pols and pol ζ is under debate. In the original paper describing the discovery, an elegant mechanism of switches of pol’s catalytic subunits on the already present core of PCNA/POLD2/POLD3 was proposed [
78], and the role of Fe-S clusters in CTDs of both pols recognized [
90,
91], but possible details of the process have never been elaborated. The argument against the switch mechanism is the stability of multi-subunit complexes of pols δ and ζ [
87,
88]. Pols’ architecture with shared subunits might reflect evolutionary relationships and structural requirements [
66].
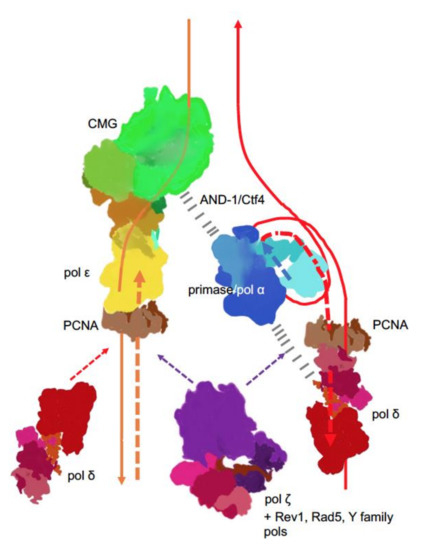
New findings lead to a better understanding of replication fork in eukaryotes (). The CMG complex bound to the C-terminal part of pol ε travels along on the leading strand. This tight association explains the participation of pol ε in the leading strand synthesis and exclusion of this pol from synthesis and proofreading on the lagging strand. In yeast pol, the accessory subunits of Pol2, Dpb3/Dpb4, may serve as “staples” rigidly connecting the C-terminal part with the active
N-terminal part [
65]. However, if this rigidity were stable, any transactions by other DNA pols on the leading strand (for example, when the switch to translesion pol is necessary for DNA damage bypass) would have been blocked by the
N-terminal half of Pol2 stuck with the primer terminus, but this is not the case. Pol δ proofreads errors made by pol ε [
25,
44] and pol ζ, with other translesion DNA synthesis pols, operate on the leading strand to the same extent as on the lagging strand [
44,
92]. Therefore, there should be a mechanism of how the active part of the catalytic subunit of pol ε abandons the 3’-end of the nascent leading strand and yields to other pols.
Figure 4. Main players at the eukaryotic replication fork. The CMG complex unwinds DNA, primase-Pol α synthesizes short RNA-DNA primers that are extended by pol δ. On the lagging strand, pol δ synthesis is halted when the pol reaches the previous Okazaki fragment. On the leading strand, pol ε takes over and contributes to around 80% of the bulk strand synthesis. Pol δ occasionally proofreads errors made by pol ε [
25] and likely continues synthesis on the leading strand thereafter. The coordination of the whole process is likely achieved by interactions of primase-pol α with CMG via Ctf4 (yeast) or AND-1 (humans) [
93,
94,
95,
96] and some not precisely mapped pol α interactions with pol δ (dashed black lines) [
97,
98,
99]. Replication stress caused by unusual DNA structures [
100], DNA damage or defects in replisome [
101,
102,
103,
104] lead to recruitment and patches of synthesis of the fourth member of the B-family, pol ζ [
105], along with translesion pols and accessory factors to mitigate replication problems, depending on the nature of replication problems.
The lagging DNA strand is synthesized in relatively short Okazaki fragments whose size coincides with the nucleosomal repeat (165 bp), as measured under conditions of constrained ligation [
106,
107]. The evidence from the distribution of inaccurate pol α-dependent mutations in yeast seems to support this estimate [
108]. The need for the ligation of short DNA fragments on the lagging strand led to a straightforward assumption that nicks and ssDNA regions are more prevalent in this strand. Such a property of the lagging strand would explain the more efficient operation of MMR on the lagging strand [
109], or preferential damage of the lagging strand by DNA editing cytosine deaminases of the APOBEC family that act on ssDNA, shown in model systems [
110,
111] and tumors [
112,
113]. However, recent findings suggest that the leading DNA strand is discontinuous as well, and nicks in the yeast’s leading strand are even more frequent than in the lagging strand [
107]. The effect is attributed to ribonucleotide excision, as seen in bacteria [
114,
115] and the preferential incorporation and repair of ribonucleotides in the leading DNA strand in yeast.
 ) is present in each of the four pol complexes.
) is present in each of the four pol complexes.