Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Allergy
Oncogenic fusion proteins, arising from chromosomal rearrangements, have emerged as prominent drivers of tumorigenesis and crucial therapeutic targets in cancer research. The detection of gene fusions is essential for the identification of cancer and the development of targeted therapeutic approaches.
- fusion proteins
- inhibitor
- therapeutic
- cancer
1. Introduction
Genomic instability is a prominent factor in driving tumorigenesis [1]. Specifically, gene fusions represent significant genomic events in the progression of cancer [2]. Gene fusions result from the physical juxtaposition of two or more previously separate genes, generating a novel chimeric gene that comprises the head of one gene and the tail of another. These chimeric genes can be generated through several possible processes, including chromosomal inversions, tandem duplications, interstitial deletions, or translocation events [3]. Gene fusions often encode fusion proteins that possess unique functional properties distinct from the original proteins, thereby leading to alterations in cellular functions and promoting tumorigenesis. Gene fusions are frequently identified as primary tumor predisposing events in leukemias, lymphomas, solid malignancies, and benign tumors [4]. Various fusion proteins have emerged as critical biomarkers for clinical diagnosis and therapeutic targets [5]. Therefore, gene fusions have garnered substantial attention within the realm of cancer research.
The discovery of the BCR–ABL fusion protein in patients with chronic myeloid leukemia (CML) marked the inception of a new era in cancer research [6]. Subsequently, numerous gene fusions have been identified in various cancer types [7], with advancements in detection methods. The application of RNA sequencing technology has revealed a multitude of unique fusion events, surpassing a count of 2200, with 120 of them exhibiting high frequency. Among these, a substantial proportion encodes protein kinases, which are known to play crucial roles in multiple signaling pathways, with relatively high expression levels and kinase activity [8]. Fusion proteins have been recognized as pivotal drivers of cancer initiation and progression in a variety of malignancies. For instance, the BCR–ABL fusion protein has been shown to induce the transformation of benign cells into malignant ones [9]. In fibrolamellar hepatocellular carcinoma (FL-HCC), the DNAJB1–PRKACA fusion drives tumorigenesis by hyperactivating the Wnt signaling pathway [10]. RET/PTC have been observed in papillary carcinomas and shown to activate the RAS-MAPK pathway in a ligand-independent way [11]. Furthermore, MAN2A1–FER has been identified in various tumor types and has been demonstrated to enhance the FER kinase activation [12]. An increasing number of studies have illustrated the pivotal role of kinase fusion proteins in the pathogenesis of cancer by aberrantly activating downstream signaling pathways. Notably, a mounting body of research has also highlighted the significance of non-kinase fusion proteins, particularly transcription factor-related non-kinase fusion proteins, in cancer development. For instance, PML–RARα and FUS–DDIT3 have been considered as the key drivers of acute myeloid leukemia (AML) and mucinous liposarcoma (MLS) [13,14]. Therefore, comprehending the mechanistic and evolutionary processes underlying fusion proteins is crucial for understanding their role in driving carcinogenesis.
The remarkable specificity exhibited by fusion genes towards cancer cells has propelled their emergence as diagnostic markers and promising targets for cancer therapies. By targeting fusion proteins, the potential arises to mitigate the off-target effects commonly associated with traditional cancer treatments, such as chemotherapy and radiation therapy, which frequently lead to many side effects and adversely impact the patients’ quality of life. Consequently, the development of selective inhibitors for fusion proteins has become a critical goal in the realm of related-cancers therapies. Targeted therapies, particularly tyrosine kinase inhibitors (TKIs), have demonstrated remarkably efficacy in treating specific cancer types, including the first-, second- and third-generation TKIs. For example, Imatinib, a drug designed to target BCR–ABL, has exhibited effectiveness in the treatment of CML [15]. Recently approved drugs such as Larotrectinib and Entrectinib are capable of targeting fusion proteins harboring neurotrophic RTK (NRTK) genes [16]. Crizotinib has demonstrated efficient inhibition of ALK fusions [17]. Additionally, all-trans-retinoic acid (ATRA) and arsenic trioxide (ATO) can be employed to treat or alleviate the blockage of granulocyte differentiation caused by the RARA fusion protein [18]. Despite the success achieved by inhibitors in the treatment of fusion-driven tumors, resistance often emerges as a result of secondary mutations [19]. Remarkably, the therapeutic strategy for cancers has undergone a paradigm shift with the advent of molecular-targeted therapy for oncogenes-driven cancers.
2. Fusion Genes in Diseases and the Detection of Fusion Genes
At the genomic level, the process of chromosome breakage and subsequent re-splicing gives rise to gene fusions, which are novel genes formed through this mechanism [21]. Unlike other recurrent oncogenic mutations, gene fusions are often regarded as pathognomonic factors for malignancy, and many of them are strongly associated with specific cancer types, such as prostate cancer [22,23], lung cancer [24,25], breast cancer, and so on.
Thus, the detection of gene fusions is essential for the identification of cancer and the development of targeted therapeutic approaches. These gene fusions generate novel chimeric proteins that can drive oncogenic transformation. The identification of gene fusions in cancer patients can provide valuable insight for personalized treatment strategies that target the aberrant fusion proteins or downstream signaling pathways, ultimately leading to improved therapeutic outcomes. Currently, a variety of techniques are wildly employed in clinical practice, include cell morphology, exon array analysis (EAA), immunohistochemistry (IHC), fluorescence in situ hybridization (FISH), reverse transcription polymerase chain reaction (RT-PCR), droplet digital PCR (DD PCR), and next-generation sequencing (NGS) (Figure 1).
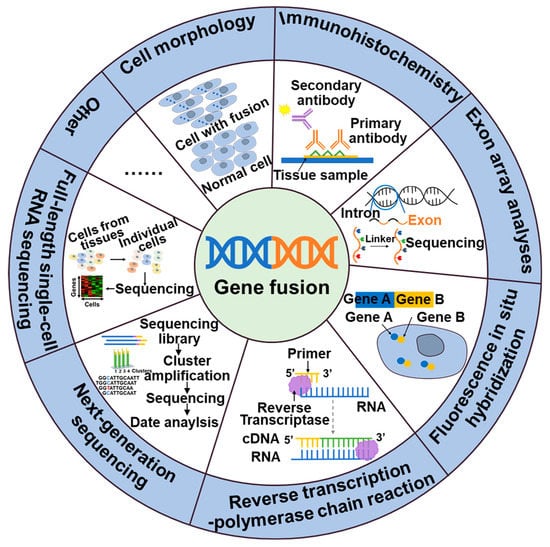
Figure 2. Methods of detection of gene fusions.
NTRK, ALK, NTRK1, BRAF, and RET fusions and many other gene fusions have previously been successfully detected by cytomorphological methods [26,27]. However, it may not always be sensitive enough to detect low-level fusion transcriptions or may not be able to differentiate between different fusion variants. The convenience and speed offered by IHC, the specificity provided by FISH, and the high detection sensitivity of RT-PCR have positioned them as widely utilized detection methods. These techniques have made great contributions to the identification of gene fusions, including ALK, NTRK, and RET. While IHC stands out for its cost-effectiveness and rapidity compared to other available assays, caution is warranted due to its tendency to yield false-positive results. Consequently, validation in combination with complementary techniques is often necessary to ensure reliable outcomes [28]. The emergence of NGS technology has facilitated targeted therapy for tumors by enabling the detection not only of fusions but also polygenes and fusion mutants [29]. In addition to the classical detection techniques, a large number of new assays are emerging, including targeted RNA sequencing (RNAseq) and full-length single-cell sequencing (scRNA-seq). Compared to FISH and RT-PCR methods, targeted RNAseq improves diagnostic yield by evaluating multiple genes simultaneously in a single assay, thereby increasing sequencing coverage and reducing diagnostic time. Additionally, it can also identify new gene fusions or complex structural rearrangements. However, the broader scope of detection in targeted RNAseq may lead to an increased likelihood of false-positive results [30]. The scRNA-seq [31], employing the detection algorithm scFusion, presents the opportunity to detect fusion genes at the single-cell level, thereby enhancing the detection power through co-analysis of relevant cells. However, the ability to detect rare fusions in highly heterogeneous tumor samples is limited. Furthermore, scRNAseq data often exhibit high levels of noise and contain various technical artifacts, which can also contribute to false-positive results [31].
This entry is adapted from the peer-reviewed paper 10.3390/molecules28124672
This entry is offline, you can click here to edit this entry!
