Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Drawing from an extensive review of 66 publications, a comprehensive overview of the diverse approaches to creating mHealth-based biomarkers using machine learning is presented herein. By exploring the current landscape of biomarker development using mHealth technologies and machine learning, researchers aim to provide valuable insights into this rapidly evolving field. By doing so, researchers reflect on current challenges in this field and propose recommendations for ensuring the development of accurate, reliable, and interpretable biomarkers.
- machine learning
- biomarker
- wearables
- smartphones
- mHealth
- remote monitoring
- central nervous system
- clinical trials
1. Introduction
1.1. Motivation
Disorders that are affected by the Central Nervous System (CNS), such as Parkinson’s Disease (PD) and Alzheimer’s Disease (AD), have a significant impact on the quality of life of patients. These disorders are often progressive and chronic, making long-term monitoring essential for assessing disease progression and treatment effects. However, the current methods for monitoring disease activity are often limited by accessibility, cost, and patient compliance [1][2]. Limited accessibility to clinics or disease monitoring devices may hinder the regular and consistent monitoring of a patient’s condition, especially for patients living in remote areas or for those who have mobility limitations. Clinical trials incur costs related to personnel, infrastructure, and equipment. A qualified healthcare team, including clinical raters, physicians, and nurses, contributes to personnel costs through salaries, training, and administrative support. Trials involving specialized equipment for measuring biomarkers can significantly impact the budget due to costs associated with procurement, maintenance, calibration, and upgrades. Furthermore, infrastructure costs may increase as suitable facilities are required for data collection during patient visits and equipment storage. Patient compliance poses challenges for disease monitoring, as some methods require patients to adhere to strict protocols, collect data at specific time intervals, or perform certain tasks that can be challenging for patients to execute. Low or no compliance can lead to incomplete or unreliable monitoring results, which in turn can hinder the reliability of the assessments. Given these limitations, there is a growing interest in exploring alternative approaches to monitoring CNS disorders that can overcome these challenges. The increasing adoption of smartphones and wearables among patients and researchers offers a promising avenue for remote monitoring.
Patient-generated data from smartphones, wearables, and other remote monitoring devices can potentially complement or supplement clinical visits by providing data during evidence gaps between visits. As the promise of mobile Health (mHealth) technologies is to provide more sensitive, ecologically valid, and frequent measures of disease activity, the data collected may enable the development and validation of novel biomarkers. The development of novel ‘digital biomarkers’ using data collected from electronic Health (eHealth) and mHealth device sensors (such as accelerometers, GPS, and microphones) offers a scalable opportunity for the continuous collection of data regarding behavioral and physiological activity under free-living conditions. Previous clinical studies have demonstrated the benefits of smartphone and wearable sensors to monitor and estimate symptom severity associated with a wide range of diseases and disorders, including cardiovascular diseases [3], mood disorders [4], and neurodegenerative disorders [5][6]. These sensors can capture a range of physiological and behavioral data, including movement, heart rate, sleep, and cognitive function, providing a wealth of information that can be used to develop biomarkers for CNS disorders in particular. These longitudinal and unobtrusive measurements are highly valuable for clinical research, providing a scalable opportunity for measuring behavioral and physiological activity in real-time. However, these approaches may carry potential pitfalls as the data sourced from these devices can be large, complex, and highly variable in terms of availability, quality, and synchronicity, which can therefore complicate analysis and interpretation [7][8]. Machine Learning (ML) may provide a solution to processing heterogenous and large datasets, identifying meaningful patterns within the datasets, and predicting complex clinical outcomes from the data. However, the complexities involved in developing biomarkers using these new technologies need to be addressed. While these tools can aid the discovery of novel and important digital biomarkers, the lack of standardization, validation, and transparency of the ML pipelines used can pose challenges for clinical, scientific, and regulatory committees.
1.2. What Is Machine Learning
In clinical research, one of the primary objectives is to understand the relationship between a set of observable variables (features) and one or more outcomes. Building a statistical model that captures the relationship between these variables and the corresponding outputs facilitates the attainment of this understanding [9]. Once this model is built, it can be used to predict the value of an output based on the features.
ML is a powerful tool for clinical research as it can be used to build statistical models. A ML model consists of a set of tunable parameters and a ML algorithm that enables the generation of outputs based on given inputs and selected parameters. Although ML algorithms are fundamentally statistical learning algorithms, ML and traditional statistical learning algorithms can differ in their objectives. Traditional statistical learning aims to create a statistical model that represents causal inference from a sample, while ML aims to build generalizable predictive models that can be used to make accurate predictions on previously unseen data [10][11]. However, it is essential to recognize that while ML models can identify relationships between variables and outcomes, they may not necessarily identify a causal link between them. This is because even though these models may achieve good performances, it is crucial to ensure that their predictions are based on relevant features rather than spurious correlations. This enables the researchers to gain meaningful insights from ML models while also being aware of their inherent limitations.
While ML is not a substitute for the clinical evaluation of patients, it can provide valuable insights into a patient’s clinical profile. ML can help to identify relevant features that clinicians may not have considered, leading to better diagnosis, treatment, and patient outcomes. Additionally, ML can help to avoid common pitfalls observed in clinical decision making by removing bias, reducing human error, and improving the accuracy of predictions [12][13][14][15]. As the volume of data generated for clinical trials and outside clinical settings continues to grow, ML’s support in processing data and informing the decision-making process becomes necessary. ML can help to uncover insights from large and complex datasets that would be difficult or impossible to identify manually.
To develop an effective ML model, it is necessary to follow a rigorous and standardized procedure. This is where ML pipelines come in. Table 1 showcases an exemplary ML pipeline, which serves as a systematic framework for automating and standardizing the model generation process. The pipeline encompasses multiple stages to ensure an organized and efficient approach to model development. First, defining the study objective guides the subsequent stages and ensures the final model meets the desired goals. Second, raw data must be preprocessed to remove errors, inconsistencies, missing data, or outliers. Third, feature extraction and selection identifies quantifiable characteristics of the data relevant to the study objective and extracts them for use in the ML model. Fourth, ML algorithms are applied to learn patterns and relationships between features, with optimal configurations identified through iterative processes until desired performance metrics are achieved. Finally, the model is validated against a new dataset that is not used in training to ensure generalizability. Effective reporting and assessment of ML procedures must be established to ensure transparency, reliability, and reproducibility.
Table 1. Representation of a standard machine learning pipeline.
| Stage | Objective | Example |
|---|---|---|
|
The ML pipeline is provided with a study objective in which the features and corresponding outputs are defined. The ML model aims to identify the associations between the features and outputs. |
The study objective is to classify Parkinson’s Disease patients and control groups using smartphone-based features. |
|
Data preprocessing filters and transforms raw data to guarantee or enhance the ML training process. | To improve the model performance, one may identify and exclude any missing or outlier data. |
|
Feature engineering uses raw data to create new features that are not readily available in the dataset. Feature selection selects the most relevant features for the model objective by removing redundant or noisy features. Together, the goal is to simplify and accelerate the computational process while also improving the model process. For deep learning methods, the concept of “feature engineering” is typically embedded within the model architecture and training process, although substantial preprocessing steps may occur prior to that. |
An interaction of two or more predictors (such as a ratio or product) or re-representation of a predictor are examples of feature engineering. Removing highly correlated or non-informative features are examples of feature selection. Note: The feature selection step can occur during model training |
|
During training, the ML model(s) iterates through all the examples in the training dataset and optimizes the parameters of the mathematical function to minimize the prediction error. To evaluate the performance of the trained ML model, the predictions of an unseen test set are compared with a known ground truth label. |
Cross-validation can be used to optimize and evaluate model performance. Classification models may be evaluated based on their prediction accuracy, sensitivity, and specificity, while regression models may be evaluated using variance explained (R2) and Mean Absolute Error. |
2. Machine Learning Algorithms
ML algorithms build a statistical model based on a training dataset, which can subsequently be used to make predictions about a new, unseen dataset. ML algorithms have been used in a wide variety of clinical trial applications, such as the classification of a diagnoses, classification of physical or mental state (such as a seizure or mood), and the estimation of symptom severity. Within the realm of clinical research, ML algorithms can be broadly divided into two learning paradigms: supervised and unsupervised learning [16].
Supervised ML algorithms use labeled data to map the patterns within a dataset to a known label, while unsupervised ML algorithms do not [17]. Rather, the unsupervised ML algorithms learn the structure present within a dataset without relying on annotations. Supervised learning can be used to automate the labelling process, detect disease cases, or predict clinical outcomes (such as treatment outcomes). There are scenarios when experts or participants can provide labelled data; however, it can become labor-intensive or time-consuming to label every observation. For example, a supervised learning algorithm trained to classify human sounds can be used to automatically annotate and quantify hours of coughs [18] and instances of crying [19]. These algorithms can also be used to differentiate between clinical populations and control participants [20] to identify known clinical population subtypes [21] or classify a clinical event (such as a seizure or tremor) [22]. The majority of the selected studies (N = 38) used a clinician to provide the label data. Some studies (N = 22) used a combination of a clinician and self-reported label data, and six studies solely relied on self-reported assessments. Unsupervised ML algorithms can be used to investigate the similarities and differences within a dataset without human intervention. This makes it the ideal solution for exploratory data analysis, subgroup phenotype identification, and anomaly detection. Among digital phenotyping studies, unsupervised learning has been used to identify location patterns [23] and classify sleep disturbance subtypes using wrist-worn accelerometer data [24].
It is important to recognize that unsupervised and supervised methods are not mutually exclusive, and they can be effectively combined. For instance, unsupervised methods can be employed to extract a meaningful latent representation of the input data. Subsequently, these latent vectors, along with the original inputs, can be used as inputs for a supervised model. This type of approach is commonly observed when applying techniques such as PCA, clustering, or other dimensionality reduction methods [25][26][27][28]. By combining unsupervised and supervised methods, valuable information can be extracted from the data and used to enhance the performance and interpretability of the overall model.
In clinical research, supervised ML algorithms have been used to classify class labels or estimate scores. Classification algorithms learn to map a new observation to a pre-defined class label. These algorithms can be used to classify patient populations and patient population subtypes and identify clinical events. Regression algorithms learn to map an observation to a continuous output. These algorithms are commonly used to estimate symptom severity [29], quantify physical activity, and forecast future events [30]. Among the selected papers that were focused on the classification of a diagnosis or state, the four most common algorithms were Random Forest, Support Vector Machine, Logistic Regression, and k-Nearest Neighbors (Figure 1). Some additional classification algorithm families identified were Naïve Bayes, Ensemble-based methods (including Decision Trees, Bagging, and Gradient Boosting), and Neural Networks (such as Convolutional, Artificial, and Recurring Neural Networks). The three most common algorithms for the regression-focused papers were Linear Regression (including linear mixed effects models), Support Vector Machine, and k-Nearest Neighbors (Figure 1). Most studies only considered or reported a single ML algorithm (N = 32). Additionally, 29 of the studies considered or reported two to five ML algorithms, and the remaining 5 studies considered six or more. The following section provides an overview of the most widely used machine learning models, their properties, advantages, and disadvantages. In addition, some notable off-the-shelf ML approaches and some custom-built ML methods such as transfer learning, multi-task learning, and generalized and personalized models, are also discussed.
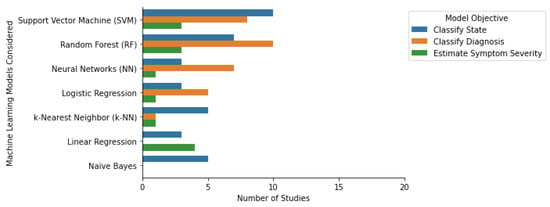
Figure 1. Machine learning algorithms and their respective objectives in the selected studies.
2.1. Tree-Based Models
A Decision Tree (DT) is a supervised non-parametric algorithm that is used for both classification and regression. A DT algorithm has a hierarchical structure in which each node represents a test of a feature, each branch represents the result of that test, and each leaf represents the class label or class distribution [31][32]. A Random Forest (RF) algorithm is a supervised ensemble learning algorithm consisting of multiple DTs that aims to predict a class or value [33]. Ensemble learning algorithms use multiple ML algorithms to obtain a prediction [34]. Tree-based models have several benefits. As each tree is only based on a subset of features and data and because they make no assumptions about the relationship between the features and distribution, they are not sensitive to collinearity between features, can ignore missing data, and are less susceptible to overfitting (for multiple trees), making the model more generalizable [35]. Another advantage of RF and DT models is that they can support linear and nonlinear relationships between the dependent and independent variables [36]. Further, as the design of the RF models can be interpreted in terms of feature importance and proximity plots, the interpretability of the RF model is feasible. However, a limitation of using tree-based models is that small changes in the data can lead to drastically different models. Additionally, the more complicated a tree-based model becomes, the less explainable a model becomes. However, pruning the trees can help to reduce the complexity of the model.
According to the selected studies, RF is a versatile and powerful model used for classification and regression tasks across multiple datatypes and populations. RF models have been used for the classification of diagnoses among PD patients [37][38], Multiple Sclerosis [39][40], and BD and unipolar depressed patients [41][42]. It is also a popular classification model for the classification of states or episodes, such as the detection of flares among Rheumatoid Arthritis or Axial Spondylarthritis patients [43] and tremor detection among PD patients [44], to quantify physical activity among cerebral palsy patients [45] and detect the moods of BD patients [46][47]. RF regression algorithms have also been used to predict anxiety deterioration among patients who suffer with anxiety [48].
2.2. Support Vector Machines
A Support Vector Machine (SVM) is a supervised algorithm that is used for classification and regression tasks. The objective of a SVM is to identify the optimal hyperplane based on the individual observations, also known as the support vectors. For SVM regression, the optimal hyperplane represents the minimal distance between the hyperplane and the support vectors. Whereas for SVM classification, the objective is to find the hyperplane that represents the maximum distance between two classes [49]. The hyperplanes can separate the classes in either a linear or non-linear fashion [36]. Given that SVMs are influenced by the support vectors closest to the hyperplanes, SVMs are less influenced by outliers, making them more suitable for extreme case binary classification. The performance of a SVM can be relatively poor when the classes are overlapping or do not have clear decision boundaries. This makes SVMs less appealing for classification tasks as inter class similarity is low. SVMs are computationally demanding models as they compute the distance between each support vector; hence, SVMs do not scale well for large datasets [50].
SVM classifiers have been used to classify clinical populations (e.g., facial nerve palsy and their control participants) [51]. SVM classifiers have also been used to classify events or states, such as detecting gait among PD patients [52] and classifying seizures among epileptic children [53]. Researchers identified studies that used SVM regression to estimate motor fluctuations and gait speed among PD and Multiple Sclerosis patients, respectively [27][54].
2.3. k-Nearest Neighbors
A k-Nearest Neighbor (k-NN) algorithm is a non-parametric supervised learning approach that can be used for multi-class classification and regression tasks. Classification k-NN algorithms determine class membership by the plurality vote of its nearest neighbors. They can estimate the continuous value of an output by calculating the average value of its nearest neighbors [36]. Given this, the quality of predictions is not only dependent on the amount of data but also on the density of the data (the number of points per unit). K-NN is simple to implement, intuitive to understand, and robust to noisy training data. However, the disadvantage is that k-NN is computationally slow when it is faced with large multi-dimensional datasets. Further, k-NN does not work well with imbalanced datasets, as under- or over-represented datapoints will influence the classification [55].
The most popular application for k-NN algorithms is for wearable-based time series data. K-NN classification models have been used to classify PD and healthy controls [56], classify tremor severity [57], predict acute exacerbations of chronic obstructive pulmonary disease (AECOPD) [58], and identify mood stability among BD and MDD patients [46][59][60]. Using wearable data, k-NN regression models have been used to predict the deterioration of symptoms associated with anxiety disorder [48].
2.4. Naïve Bayes
A Naïve Bayes (NB) classifier is a supervised multi-class classification algorithm. NB classifiers calculate the class conditional probability—the probability that a datapoint belongs to a given class in the data [49][61]. NB classifiers are computational efficient algorithms; thus, they are suitable for real-time predictions, scale well for larger datasets, and can handle missing values. A limitation of NB is that it assumes that all features are conditionally independent; hence, it is recommended that collinear features are removed in advance. Another limitation is that when new feature-observation pairs do not resemble the data in the training data, the NB assigns a probability of zero to that observation. This approach is particularly harsh, especially when dealing with a smaller dataset. Hence, the training data should represent the entire population.
As NB classifiers help form classification models, it is found that NB classifiers have been used for the classification of tremors or for freezing gait among PD patients [62], as well as to classify flares among Rheumatoid Arthritis and Axial Spondylarthritis patients [43] and classify bipolar episodes and mood stability among BD and MDD patients [46][59][60].
2.5. Linear and Logistic Regression
A Linear Regression model is a supervised regression model that predicts a continuous output. It finds the optimal hyperplane that minimizes the sum of squared difference between the true data points and the hyperplane. A Logistic Regression model is a supervised classification model that can be used for binomial, multinominal, and ordinal classification tasks. Logistic Regression classifies observations by examining the outcome variables on the extreme ends and determines a logistic line that divides two or more classes [36]. Linear and Logistic Regression are popular in algorithms as they are easy to implement, efficient to train, and easy to interpret. However, a limitation of both models is that they make multiple assumptions, e.g., that a solution is linear, the input residuals are normally distributed, and that all features are mutually independent [63]. Multicollinearity, the correlation between multiple features, and outliers will inflate the standard error of the model and may undermine the significance of significant features [64]. Further, outliers that deviate from the expected range of the data can skew the extreme bounds of the probability, making both algorithms sensitive to outliers in the dataset [63].
Linear Regression has been used to quantify tremors among Essential Tremor (ET) patients [65] and to estimate motor-related symptom severity among PD patients [66][67]. It has also been used to forecast convergence between body sides for Hemiparetic patients [30]. Logistic Regression was a popular approach for classifying PD diagnosis [37][38], Post-Traumatic Stress Disorder [68], and distinguishing fallers and non-fallers [69]. Logistic Regression has been used to classify drug effects, such as predicting the pre- and post-medication states among PD patients [70].
2.6. Neural Networks
Neural Networks (NN), also known as Artificial Neural Networks (ANN), can be used for unsupervised and supervised classification and regression tasks [71]. NN consists of a collection of artificial neurons (or nodes). Each artificial neuron receives, processes, and sends the signal to the artificial neuron connected to it. The neurons are aggregated into multiple layers, and each layer performs different transformations on the signal. The signal first travels from the input layer into the output layer while possibly traversing multiple hidden layers in between. NN offer several advantages, such as the ability to detect complex non-linear relationships between features and outcomes and work with missing data, while it also requires less preprocessing of the data and offers the availability of multiple training algorithms. However, the disadvantages of NN include increased computational burden, reduced explainability and interpretability (as NN are ‘black box’ in nature), and the fact that NN are prone to overfitting [72]. However, it is important to highlight the growing number of studies that specifically explore explainable deep learning approaches for biomarker discovery and development. Studies utilizing methodologies such as LIME (Lime Tabular Explainer), SHAP (Shapley Additive exPlanations), and other visual inspections of feature distribution and importance have aided clinicians in understanding the model mechanisms. These approaches also provide patient-specific insights by describing the importance of each feature, which may, in turn, facilitate personalized treatment opportunities [73][74][75][76].
The most popular applications for neural networks were for the classification of a diagnosis or classification of a state or event. The most popular application is the detection of tremors among PD patients [21][44][62][77][78]. NN have been used to classify unipolar and bipolar depressed patients based on motor activity [41][79], estimate depression severity [79], forecast seizures [80], and classify a treatment response using keyboard patterns among PD patients [81].
2.7. Transfer Learning
Transfer learning (also known as domain adaption) refers to the act of deriving the representations of a previously trained ML model to extract meaningful features from another dataset for an inter-related task [82]. One applicable scenario is the training of a supervised ML model on data collected in a controlled setting (such as in a lab or clinic). The performance of the model may suffer when applied to a dataset collected under free-living conditions. Rather than developing a new model trained solely on a free-living condition dataset, transfer learning can use patterns learned from the controlled setting dataset to improve the learning of the patterns from the free-living conditions dataset.
Transfer learning can also be a valuable technique for enhancing the utilization of limited or rare data [83]. One practical application is to employ pretraining on abundant control data and subsequently finetune the model on the specific population of interest to improve the model’s performance [83][84][85]. This approach not only optimizes the efficiency of utilizing scarce data but also facilitates model personalization. By adapting a pretrained model to individual characteristics or preferences, it becomes possible to create personalized models that better cater to unique needs or circumstances. Transfer learning thus offers a powerful means to leverage existing knowledge and make the most of available data resources, enhancing both the efficiency and personalization of biomarkers.
Given its application, transfer learning reduces the amount of labeled data and computational resources required to train new ML models [82], thus making this method advantageous when the sensor modalities, sensor placements, and populations differ between studies. While researchers only identified two studies that applied transfer learning to estimate PD disease severity using movement sensor data [86][87], researchers predict that the application of transfer learning will enable future researchers to overcome the challenges of a limited dataset and develop more sensitive and effective ML models.
2.8. Multi-Task Learning
Multi-task learning (MTL) enables the learning of multiple tasks simultaneously [88]. Learning the commonalities and differences between multiple tasks can improve both the learning efficiency and the prediction accuracy of the ML models [88]. A traditional single-task ML model can have a performance ceiling effect, given the limitations of the dataset size and the model’s ability to learn meaningful representations. MTL uses all available data across multiple datasets and can learn to develop generalized models that are applicable to multiple tasks. To use MTL, there should be some degree of information shared between or across all tasks. The correlation allows MTL to exploit the underlying shared information or principles within tasks. Sometimes MTL models can perform worse than single-task models because of ‘negative transfers’. This occurs when different tasks share no mutual information or if the information of tasks are contradictory [89]. MTL models have been used to simultaneously model data sourced from two separate sources or to model multiple outcomes [90][91]. For example, Lu et al. explored the use of MTL to jointly model data collected from two different smartphone platforms (iPhone and Android) to jointly predict two different types of depression assessments (QIDS and a DSM-5 survey) [92]. They illustrated that the classification accuracy of the MTL approach outperformed the single-task learning approach by 48%; thus, the classification model benefited from learning from observations sourced from multiple devices.
2.9. Generalized vs. Personalized
ML algorithms can be trained on population data or individual subject data. Generalized models, which are trained on population data, are fed data from all participants for the purpose of general knowledge learning. Conversely, personalized models are trained on an individual’s data and take into consideration individual factors such as biological or lifestyle-related variations [93]. The researchers have adopted these terms from Kahdemi et al.’s study, in which they developed generalized and personalized models for sleep-wake prediction [94]. The heterogenous nature of each population or individual can be a potential hinderance for generalizable models. A single individual’s deviation from the ‘norm’ may be viewed as a source of ‘noise’ in a generalized model. For example, patients with mood disorders such as MDD and BD have large inter-individual symptom variability. Abdullah et al., reliably predicted the social rhythms of BD patients with personalized models using smartphone activity data [95]. Cho et al. compared the mood prediction accuracy of personalized and generalized models based on the circadian rhythms of MDD and BD participants [96]. Their studies illustrated that their personalized model predictions were, on average, 24% more accurate than the generalized models. These studies lay the groundwork for developing personalized models that are more sensitive to individual differences.
This entry is adapted from the peer-reviewed paper 10.3390/s23115243
References
- Au, R.; Lin, H.; Kolachalama, V.B. Tele-Trials, Remote Monitoring, and Trial Technology for Alzheimer’s Disease Clinical Trials. In Alzheimer’s Disease Drug Development; Cambridge University Press: Cambridge, UK, 2022; pp. 292–300.
- Inan, O.T.; Tenaerts, P.; Prindiville, S.A.; Reynolds, H.R.; Dizon, D.S.; Cooper-Arnold, K.; Turakhia, M.; Pletcher, M.J.; Preston, K.L.; Krumholz, H.M.; et al. Digitizing clinical trials. NPJ Digit. Med. 2020, 3, 101.
- Teo, J.X.; Davila, S.; Yang, C.; Hii, A.A.; Pua, C.J.; Yap, J.; Tan, S.Y.; Sahlén, A.; Chin, C.W.-L.; Teh, B.T.; et al. Digital phenotyping by consumer wearables identifies sleep-associated markers of cardiovascular disease risk and biological aging. bioRxiv 2019.
- Brietzke, E.; Hawken, E.R.; Idzikowski, M.; Pong, J.; Kennedy, S.H.; Soares, C.N. Integrating digital phenotyping in clinical characterization of individuals with mood disorders. Neurosci. Biobehav. Rev. 2019, 104, 223–230.
- Kourtis, L.C.; Regele, O.B.; Wright, J.M.; Jones, G.B. Digital biomarkers for Alzheimer’s disease: The mobile/wearable devices opportunity. NPJ Digit. Med. 2019, 2, 9.
- Bhidayasiri, R.; Mari, Z. Digital phenotyping in Parkinson’s disease: Empowering neurologists for measurement-based care. Park. Relat. Disord. 2020, 80, 35–40.
- Prosperi, M.; Min, J.S.; Bian, J.; Modave, F. Big data hurdles in precision medicine and precision public health. BMC Med. Inform. Decis. Mak. 2018, 18, 139.
- Torres-Sospedra, J.; Ometov, A. Data from Smartphones and Wearables. Data 2021, 6, 45.
- García-Santıllán, A.; del Flóres-Serrano, S.; López-Morales, J.S.; Rios-Alvarez, L.R. Factors Associated that Explain Anxiety toward Mathematics on Undergraduate Students. (An Empirical Study in Tierra Blanca Veracruz-México). Mediterr. J. Soc. Sci. 2014, 5.
- Iniesta, R.; Stahl, D.; Mcguffin, P. Machine learning, statistical learning and the future of biological research in psychiatry. Psychol. Med. 2016, 46, 2455–2465.
- Rajula, H.S.R.; Verlato, G.; Manchia, M.; Antonucci, N.; Fanos, V. Comparison of Conventional Statistical Methods with Machine Learning in Medicine: Diagnosis, Drug Development, and Treatment. Medicina 2020, 56, 455.
- Getz, K.A.; Rafael, A.C. Trial watch: Trends in clinical trial design complexity. Nat. Rev. Drug. Discov. 2017, 16, 307.
- Getz, K.A.; Stergiopoulos, S.; Marlborough, M.; Whitehill, J.; Curran, M.; Kaitin, K.I. Quantifying the Magnitude and Cost of Collecting Extraneous Protocol Data. Am. J. Ther. 2015, 22, 117–124.
- Getz, K.A.; Wenger, J.; Campo, R.A.; Seguine, E.S.; Kaitin, K.I. Assessing the Impact of Protocol Design Changes on Clinical Trial Performance. Am. J. Ther. 2008, 15, 450–457.
- Globe Newswire. Rising Protocol Design Complexity Is Driving Rapid Growth in Clinical Trial Data Volume, According to Tufts Center for the Study of Drug Development. Available online: https://www.globenewswire.com/news-release/2021/01/12/2157143/0/en/Rising-Protocol-Design-Complexity-Is-Driving-Rapid-Growth-in-Clinical-Trial-Data-Volume-According-to-Tufts-Center-for-the-Study-of-Drug-Development.html (accessed on 12 January 2021).
- Zhang, X.D. Machine Learning. In A Matrix Algebra Approach to Artificial Intelligence; Springer: Singapore, 2020.
- Russell, S.J.; Norvig, P. Artificial Intelligence: A Modern Approach, 4th ed.; Prentice Hall: Hoboken, NJ, USA, 2020.
- Tinschert, P.; Rassouli, F.; Barata, F.; Steurer-Stey, C.; Fleisch, E.; Puhan, M.; Kowatsch, T.; Brutsche, M.H. Smartphone-Based Cough Detection Predicts Asthma Control—Description of a Novel, Scalable Digital Biomarker; European Respiratory Society (ERS): Lausanne, Switzerland, 2020; p. 4569.
- ZhuParris, A.; Kruizinga, M.D.; van Gent, M.; Dessing, E.; Exadaktylos, V.; Doll, R.J.; Stuurman, F.E.; Driessen, G.A.; Cohen, A.F. Development and Technical Validation of a Smartphone-Based Cry Detection Algorithm. Front. Pediatr. 2021, 9, 262.
- Creagh, A.P.; Simillion, C.; Bourke, A.K.; Scotland, A.; Lipsmeier, F.; Bernasconi, C.; van Beek, J.; Baker, M.; Gossens, C.; Lindemann, M.; et al. Smartphone-and Smartwatch-Based Remote Characterisation of Ambulation in Multiple Sclerosis during the Two-Minute Walk Test. IEEE J. Biomed. Health Inform. 2021, 25, 838–849.
- Papadopoulos, A.; Kyritsis, K.; Klingelhoefer, L.; Bostanjopoulou, S.; Chaudhuri, K.R.; Delopoulos, A. Detecting Parkinsonian Tremor from IMU Data Collected In-The-Wild using Deep Multiple-Instance Learning. IEEE J. Biomed. Health Inform. 2019, 24, 2559–2569.
- Fatima, M.; Pasha, M. Survey of Machine Learning Algorithms for Disease Diagnostic. J. Intell. Learn. Syst. Appl. 2017, 9, 1–16.
- Faurholt-Jepsen, M.; Busk, J.; Vinberg, M.; Christensen, E.M.; HelgaÞórarinsdóttir; Frost, M.; Bardram, J.E.; Kessing, L.V. Daily mobility patterns in patients with bipolar disorder and healthy individuals. J. Affect. Disord. 2021, 278, 413–422.
- Ensari, I.; Caceres, B.A.; Jackman, K.B.; Suero-Tejeda, N.; Shechter, A.; Odlum, M.L.; Bakken, S. Digital phenotyping of sleep patterns among heterogenous samples of Latinx adults using unsupervised learning. Sleep. Med. 2021, 85, 211–220.
- Palmius, N.; Tsanas, A.; Saunders, K.E.A.; Bilderbeck, A.C.; Geddes, J.R.; Goodwin, G.M.; De Vos, M. Detecting bipolar depression from geographic location data. IEEE Trans. Biomed. Eng. 2017, 64, 1761–1771.
- Rodríguez-Martín, D.; Samà, A.; Pérez-López, C.; Català, A.; Arostegui, J.M.M.; Cabestany, J.; Bayés, À.; Alcaine, S.; Mestre, B.; Prats, A.; et al. Home detection of freezing of gait using Support Vector Machines through a single waist-worn triaxial accelerometer. PLoS ONE 2017, 12, e0171764.
- Supratak, A.; Datta, G.; Gafson, A.R.; Nicholas, R.; Guo, Y.; Matthews, P.M. Remote monitoring in the home validates clinical gait measures for multiple sclerosis. Front. Neurol. 2018, 9, 561.
- Ko, Y.-F.; Kuo, P.-H.; Wang, C.-F.; Chen, Y.-J.; Chuang, P.-C.; Li, S.-Z.; Chen, B.-W.; Yang, F.-C.; Lo, Y.-C.; Yang, Y.; et al. Quantification Analysis of Sleep Based on Smartwatch Sensors for Parkinson’s Disease. Biosensors 2022, 12, 74.
- Farhan, A.A.; Yue, C.; Morillo, R.; Ware, S.; Lu, J.; Bi, J.; Kamath, J.; Russell, A.; Bamis, A.; Wang, B. Behavior vs. introspection: Refining prediction of clinical depression via smartphone sensing data. In Proceedings of the 2016 IEEE Wireless Health (WH), Bethesda, MD, USA, 25–27 October 2016.
- Derungs, A.; Schuster-Amft, C.; Amft, O. Longitudinal walking analysis in hemiparetic patients using wearable motion sensors: Is there convergence between body sides? Front. Bioeng. Biotechnol. 2018, 6, 57.
- Freedman, D.A. Statistical Models. In Statistical Models: THeory and Practice; Cambridge University Press: Cambridge, UK, 2009.
- Ahmed, S.T.; Basha, S.M.; Arumugam, S.R.; Kodabagi, M.M. Pattern Recognition: An Introduction, 1st ed.; MileStone Research Publications: Bengaluru, India, 2021.
- Ruppert, D. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. J. Am. Stat. Assoc. 2004, 99, 567.
- Opitz, D.; Maclin, R. Popular Ensemble Methods: An Empirical Study. J. Artif. Intell. Res. 1999, 11, 169–198.
- Kosasi, S. Perancangan Prototipe Sistem Pemesanan Makanan dan Minuman Menggunakan Mobile Device. Indones. J. Netw. Secur. 2015, 1, 1–10.
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer Science & Business Media: New York, NY, USA, 2013.
- Prince, J.; Andreotti, F.; De Vos, M. Multi-Source Ensemble Learning for the Remote Prediction of Parkinson’s Disease in the Presence of Source-Wise Missing Data. IEEE Trans. Biomed. Eng. 2019, 66, 1402–1411.
- Tracy, J.M.; Özkanca, Y.; Atkins, D.C.; Ghomi, R.H. Investigating voice as a biomarker: Deep phenotyping methods for early detection of Parkinson’s disease. J. Biomed. Inform. 2020, 104, 103362.
- Schwab, P.; Karlen, W. A Deep Learning Approach to Diagnosing Multiple Sclerosis from Smartphone Data. IEEE J. Biomed. Health Inform. 2021, 25, 1284–1291.
- Livingston, E.; Cao, J.; Dimick, J.B. Tread carefully with stepwise regression. Arch. Surg. 2010, 145, 1039–1040.
- Jakobsen, P.; Garcia-Ceja, E.; Riegler, M.; Stabell, L.A.; Nordgreen, T.; Torresen, J.; Fasmer, O.B.; Oedegaard, K.J. Applying machine learning in motor activity time series of depressed bipolar and unipolar patients compared to healthy controls. PLoS ONE 2020, 15, e0231995.
- Garcia-Ceja, E.; Riegler, M.; Jakobsen, P.; Torresen, J.; Nordgreen, T.; Oedegaard, K.J.; Fasmer, O.B. Motor Activity Based Classification of Depression in Unipolar and Bipolar Patients. In Proceedings of the 2018 IEEE 31st International Symposium on Computer-Based Medical Systems (CBMS), Karlstad, Sweden, 18–21 June 2018; pp. 316–321.
- Gossec, L.; Guyard, F.; Leroy, D.; Lafargue, T.; Seiler, M.; Jacquemin, C.; Molto, A.; Sellam, J.; Foltz, V.; Gandjbakhch, F.; et al. Detection of Flares by Decrease in Physical Activity, Collected Using Wearable Activity Trackers in Rheumatoid Arthritis or Axial Spondyloarthritis: An Application of Machine Learning Analyses in Rheumatology. Arthritis Care Res. 2019, 71, 1336–1343.
- San-Segundo, R.; Zhang, A.; Cebulla, A.; Panev, S.; Tabor, G.; Stebbins, K.; Massa, R.E.; Whitford, A.; de la Torre, F.; Hodgins, J. Parkinson’s disease tremor detection in the wild using wearable accelerometers. Sensors 2020, 20, 5817.
- Ahmadi, M.N.; O’neil, M.E.; Baque, E.; Boyd, R.N.; Trost, S.G. Machine learning to quantify physical activity in children with cerebral palsy: Comparison of group, group-personalized, and fully-personalized activity classification models. Sensors 2020, 20, 3976.
- Maxhuni, A.; Muñoz-Meléndez, A.; Osmani, V.; Perez, H.; Mayora, O.; Morales, E.F. Classification of bipolar disorder episodes based on analysis of voice and motor activity of patients. Pervasive Mob. Comput. 2016, 31, 50–66.
- Faurholt-Jepsen, M.; Busk, J.; Frost, M.; Vinberg, M.; Christensen, E.M.; Winther, O.; Bardram, J.E.; Kessing, L.V. Voice analysis as an objective state marker in bipolar disorder. Transl. Psychiatry 2016, 6, e856.
- Jacobson, N.C.; Lekkas, D.; Huang, R.; Thomas, N. Deep learning paired with wearable passive sensing data predicts deterioration in anxiety disorder symptoms across 17–18 years. J. Affect. Disord. 2021, 282, 104–111.
- Hastie, T.; Tibshirani, R.; Friedman, J. Statistics the Elements of Statistical Learning. Math. Intell. 2009, 27, 83–85.
- Patle, A.; Chouhan, D.S. SVM kernel functions for classification. In Proceedings of the 2013 International Conference on Advances in Technology and Engineering, ICATE 2013, Mumbai, India, 23–25 January 2013.
- Kim, H.S.; Kim, S.Y.; Kim, Y.H.; Park, K.S. A smartphone-based automatic diagnosis system for facial nerve palsy. Sensors 2015, 15, 26756–26768.
- Rodriguez-Molinero, A.; Samà, A.; Pérez-Martínez, D.A.; López, C.P.; Romagosa, J.; Bayes, A.; Sanz, P.; Calopa, M.; Gálvez-Barrón, C.; De Mingo, E.; et al. Validation of a portable device for mapping motor and gait disturbances in Parkinson’s disease. JMIR Mhealth Uhealth 2015, 3, e9.
- Luca, S.; Karsmakers, P.; Cuppens, K.; Croonenborghs, T.; Van de Vel, A.; Ceulemans, B.; Lagae, L.; Van Huffel, S.; Vanrumste, B. Detecting rare events using extreme value statistics applied to epileptic convulsions in children. Artif. Intell. Med. 2014, 60, 89–96.
- Ghoraani, B.; Hssayeni, M.D.; Bruack, M.M.; Jimenez-Shahed, J. Multilevel Features for Sensor-Based Assessment of Motor Fluctuation in Parkinson’s Disease Subjects. IEEE J. Biomed. Health Inform. 2020, 24, 1284–1295.
- Kramer, O. K-Nearest Neighbors. In Dimensionality Reduction with Unsupervised Nearest Neighbors. Intelligent Systems Reference Library; Springer: Berlin/Heidelberg, Germany, 2013; Volume 51.
- Tougui, I.; Jilbab, A.; El Mhamdi, J. Analysis of smartphone recordings in time, frequency, and cepstral domains to classify Parkinson’s disease. Healthc. Inform. Res. 2020, 26, 274–283.
- Jeon, H.; Lee, W.; Park, H.; Lee, H.J.; Kim, S.K.; Kim, H.B.; Jeon, B.; Park, K.S. Automatic classification of tremor severity in Parkinson’s disease using awearable device. Sensors 2017, 17, 2067.
- Wu, C.-T.; Li, G.-H.; Huang, C.-T.; Cheng, Y.-C.; Chen, C.-H.; Chien, J.-Y.; Kuo, P.-H.; Kuo, L.-C.; Lai, F. Acute exacerbation of a chronic obstructive pulmonary disease prediction system using wearable device data, machine learning, and deep learning: Development and cohort study. JMIR Mhealth Uhealth 2021, 9, e22591.
- Bai, R.; Xiao, L.; Guo, Y.; Zhu, X.; Li, N.; Wang, Y.; Chen, Q.; Feng, L.; Wang, Y.; Yu, X.; et al. Tracking and monitoring mood stability of patients with major depressive disorder by machine learning models using passive digital data: Prospective naturalistic multicenter study. JMIR Mhealth Uhealth 2021, 9, e24365.
- Grunerbl, A.; Muaremi, A.; Osmani, V.; Bahle, G.; Ohler, S.; Troster, G.; Mayora, O.; Haring, C.; Lukowicz, P. Smartphone-based recognition of states and state changes in bipolar disorder patients. IEEE J. Biomed. Health Inform. 2015, 19, 140–148.
- Pranckevičius, T.; Marcinkevičius, V. Comparison of Naive Bayes, Random Forest, Decision Tree, Support Vector Machines, and Logistic Regression Classifiers for Text Reviews Classification. Balt. J. Mod. Comput. 2017, 5, 221–232.
- Bazgir, O.; Habibi, S.A.H.; Palma, L.; Pierleoni, P.; Nafees, S. A classification system for assessment and home monitoring of tremor in patients with Parkinson’s disease. J. Med. Signals Sens. 2018, 8, 65–72.
- Worster, A.; Fan, J.; Ismaila, A. Understanding linear and logistic regression analyses. Can. J. Emerg. Med. 2007, 9, 111–113.
- Morrow-Howell, N. The M word: Multicollinearity in multiple regression. Soc. Work. Res. 1994, 18, 247–251.
- Pulliam, C.; Eichenseer, S.; Goetz, C.; Waln, O.; Hunter, C.; Jankovic, J.; Vaillancourt, D.; Giuffrida, J.; Heldman, D. Continuous in-home monitoring of essential tremor. Park. Relat. Disord. 2014, 20, 37–40.
- Ramsperger, R.; Meckler, S.; Heger, T.; van Uem, J.; Hucker, S.; Braatz, U.; Graessner, H.; Berg, D.; Manoli, Y.; Serrano, J.A.; et al. Continuous leg dyskinesia assessment in Parkinson’s disease -clinical validity and ecological effect. Park. Relat. Disord. 2016, 26, 41–46.
- Galperin, I.; Hillel, I.; Del Din, S.; Bekkers, E.M.; Nieuwboer, A.; Abbruzzese, G.; Avanzino, L.; Nieuwhof, F.; Bloem, B.R.; Rochester, L.; et al. Associations between daily-living physical activity and laboratory-based assessments of motor severity in patients with falls and Parkinson’s disease. Park. Relat. Disord. 2019, 62, 85–90.
- Cakmak, A.S.; Alday, E.A.P.; Da Poian, G.; Rad, A.B.; Metzler, T.J.; Neylan, T.C.; House, S.L.; Beaudoin, F.L.; An, X.; Stevens, J.S.; et al. Classification and Prediction of Post-Trauma Outcomes Related to PTSD Using Circadian Rhythm Changes Measured via Wrist-Worn Research Watch in a Large Longitudinal Cohort. IEEE J. Biomed. Health Inform. 2021, 25, 2866–2876.
- Schwenk, M.; Hauer, K.; Zieschang, T.; Englert, S.; Mohler, J.; Najafi, B. Sensor-derived physical activity parameters can predict future falls in people with dementia. Gerontology 2014, 60, 483–492.
- Evers, L.J.; Raykov, Y.P.; Krijthe, J.H.; de Lima, A.L.S.; Badawy, R.; Claes, K.; Heskes, T.M.; Little, M.A.; Meinders, M.J.; Bloem, B.R. Real-life gait performance as a digital biomarker for motor fluctuations: The validation study. J. Med. Internet Res. 2020, 22, e19068.
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444.
- Tu, J.V. Advantages and disadvantages of using artificial neural networks versus logistic regression for predicting medical outcomes. J. Clin. Epidemiol. 1996, 49, 1225–1231.
- Jabar, H.; Khan, R.Z. Methods to avoid over-fitting and under-fitting in supervised machine learning (comparative study). Comput. Sci. Commun. Instrum. Devices 2015, 70, 163–172.
- Mudiyanselage, T.K.B.; Xiao, X.; Zhang, Y.; Pan, Y. Deep Fuzzy Neural Networks for Biomarker Selection for Accurate Cancer Detection. IEEE Trans. Fuzzy Syst. 2020, 28, 3219–3228.
- Yagin, F.H.; Cicek, I.B.; Alkhateeb, A.; Yagin, B.; Colak, C.; Azzeh, M.; Akbulut, S. Explainable artificial intelligence model for identifying COVID-19 gene biomarkers. Comput. Biol. Med. 2023, 154, 106619.
- Wang, Y.; Lucas, M.; Furst, J.; Fawzi, A.A.; Raicu, D. Explainable Deep Learning for Biomarker Classification of OCT Images. In Proceedings of the 2020 IEEE 20th International Conference on Bioinformatics and Bioengineering (BIBE), Cincinnati, OH, USA, 26–28 October 2020; pp. 204–210.
- Cole, B.T.; Roy, S.H.; De Luca, C.J.; Nawab, S.H. Dynamical learning and tracking of tremor and dyskinesia from wearable sensors. IEEE Trans. Neural Syst. Rehabil. Eng. 2014, 22, 982–991.
- Fisher, J.M.; Hammerla, N.Y.; Ploetz, T.; Andras, P.; Rochester, L.; Walker, R.W. Unsupervised home monitoring of Parkinson’s disease motor symptoms using body-worn accelerometers. Park. Relat. Disord. 2016, 33, 44–50.
- Frogner, J.I.; Noori, F.M.; Halvorsen, P.; Hicks, S.A.; Garcia-Ceja, E.; Torresen, J.; Riegler, M.A. One-dimensional convolutional neural networks on motor activity measurements in detection of depression. In Proceedings of the HealthMedia 2019—Proceedings of the 4th International Workshop on Multimedia for Personal Health and Health Care, Co-Located with MM 2019, Nice, France, 21–25 October 2019; pp. 9–15.
- Meisel, C.; el Atrache, R.; Jackson, M.; Schubach, S.; Ufongene, C.; Loddenkemper, T. Machine learning from wristband sensor data for wearable, noninvasive seizure forecasting. Epilepsia 2020, 61, 2653–2666.
- Matarazzo, M.; Arroyo-Gallego, T.; Montero, P.; Puertas-Martín, V.; Butterworth, I.; Mendoza, C.S.; Ledesma-Carbayo, M.J.; Catalán, M.J.; Molina, J.A.; Bermejo-Pareja, F.; et al. Remote Monitoring of Treatment Response in Parkinson’s Disease: The Habit of Typing on a Computer. Mov. Disord. 2019, 34, 1488–1495.
- Weiss, K.; Khoshgoftaar, T.M.; Background, D.W. A survey of transfer learning. J. Big Data 2016, 3, 1345–1459.
- Kamishima, T.; Hamasaki, M.; Akaho, S. TrBagg: A Simple Transfer Learning Method and its Application to Personalization in Collaborative Tagging. In Proceedings of the 2009 Ninth IEEE International Conference on Data Mining, Miami, FL, USA, 6–9 December 2009; pp. 219–228.
- Fu, Z.; He, X.; Wang, E.; Huo, J.; Huang, J.; Wu, D. Personalized Human Activity Recognition Based on Integrated Wearable Sensor and Transfer Learning. Sensors 2021, 21, 885.
- Chen, Y.; Qin, X.; Wang, J.; Yu, C.; Gao, W. FedHealth: A Federated Transfer Learning Framework for Wearable Healthcare. IEEE Intell. Syst. 2020, 35, 83–93.
- Goschenhofer, J.; Pfister, F.M.J.; Yuksel, K.A.; Bischl, B.; Fietzek, U.; Thomas, J. Wearable-Based Parkinson’s Disease Severity Monitoring Using Deep Learning. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2020; Volume 11908 LNAI, pp. 400–415.
- Hssayeni, M.D.; Jimenez-Shahed, J.; Burack, M.A.; Ghoraani, B. Ensemble deep model for continuous estimation of Unified Parkinson’s Disease Rating Scale III. Biomed. Eng. Online 2021, 20, 1–20.
- Zhang, Y.; Yang, Q. Special Topic: Machine Learning An overview of multi-task learning. Natl. Sci. Rev. 2018, 5, 30–43.
- Lee, G.; Yang, E.; Hwang, S. Asymmetric multi-task learning based on task relatedness and loss. In Proceedings of the International Conference on Machine Learning 2016, New York, NY, USA, 19–24 June 2016; pp. 230–238.
- Xin, W.; Bi, J.; Yu, S.; Sun, J.; Song, M. Multiplicative Multitask Feature Learning. J. Mach. Learn. Res. JMLR 2016, 17, 1–33.
- Zhang, Z.; Jung, T.P.; Makeig, S.; Pi, Z.; Rao, B.D. Spatiotemporal sparse Bayesian learning with applications to compressed sensing of multichannel physiological signals. IEEE Trans. Neural Syst. Rehabil. Eng. 2014, 22, 1186–1197.
- Lu, J.; Shang, C.; Yue, C.; Morillo, R.; Ware, S.; Kamath, J.; Bamis, A.; Russell, A.; Wang, B.; Bi, J. Joint Modeling of Heterogeneous Sensing Data for Depression Assessment via Multi-task Learning. In Proceedings of the Proceedings of the ACM on Interactive, Mobile, Wearable and Ubiquitous Technologies; Association for Computing Machinery: New York, NY, USA, 2018; Volume 2, pp. 1–21.
- Schneider, J.; Vlachos, M. Personalization of deep learning. In Data Science–Analytics and Applications: Proceedings of the 3rd International Data Science Conference–iDSC2020; Springer: Wiesbaden, Geramny, 2021; pp. 89–96.
- Khademi, A.; El-Manzalawy, Y.; Buxton, O.M.; Honavar, V. Toward personalized sleep-wake prediction from actigraphy. In Proceedings of the 2018 IEEE EMBS International Conference on Biomedical and Health Informatics, BHI 2018, Vegas, NV, USA, 4–7 March 2018; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2018; pp. 414–417.
- Abdullah, S.; Matthews, M.; Frank, E.; Doherty, G.; Gay, G.; Choudhury, T. Automatic detection of social rhythms in bipolar disorder. J. Am. Med. Inform. Assoc. 2016, 23, 538–543.
- Cho, C.H.; Lee, T.; Kim, M.G.; In, H.P.; Kim, L.; Lee, H.J. Mood prediction of patients with mood disorders by machine learning using passive digital phenotypes based on the circadian rhythm: Prospective observational cohort study. J. Med. Internet Res. 2019, 21, e11029.
This entry is offline, you can click here to edit this entry!
