Ochrobactrum spp. are found in a wide range of environments including soils and water. They are non-enteric, Gram-negative organisms that are closely related to the genus Brucella. Several species of this genus have now been characterised and implicated as opportunistic pathogens in multiple outbreaks. This entry gives a brief overview of the Genus Ochrobactrum.
- Genus Ochrobactrum
- pathogens
- environmental bacteria
1. Introduction
Gram-negative, non-fermenting bacteria are an emergent worry in medical situations and are becoming a growing cause of severe infections. Pathogens of this type are opportunistic and include many different bacterial species, such as Ralstonia spp., Pseudomonas aeruginosa, Sphingomonas paucimobilis and Brevundimonas spp. [1][2][3][4][5]. Gram-negative, non-fermenting bacteria can infect both patients undergoing treatments and individuals outside of a clinical setting with various underlying conditions or diseases. Another type of these bacteria are the members of the α-proteobacterial genus Ochrobactrum [6].
Ochrobactrum spp. are found in a wide variety of environments including water, aircraft water, soil, plants and animals [6][7][8][9][10][11][12]. Several Ochrobactrum spp. have been investigated for their potential to degrade xenobiotic pollutants and for heavy metal detoxification under a variety of environmental conditions [13][14][15][16]. Ochrobactrum spp. are very closely related to brucellae, and even though they are considered to be of low virulence, they have increasingly been found to cause infections (some serious including endocarditis and septicaemia) in immunocompetent hosts [17][18].
Investigation of the scientific/medical literature presented a wide variety of infections resultant from Ochrobactrum spp. and these were resistant to wide variety of antibiotics. Our data point to the genus being a more common pathogen than previously supposed, with many of the infections/conditions caused by Ochrobactrum spp. being aggressive and debilitating.
2. Genus Ochrobactrum
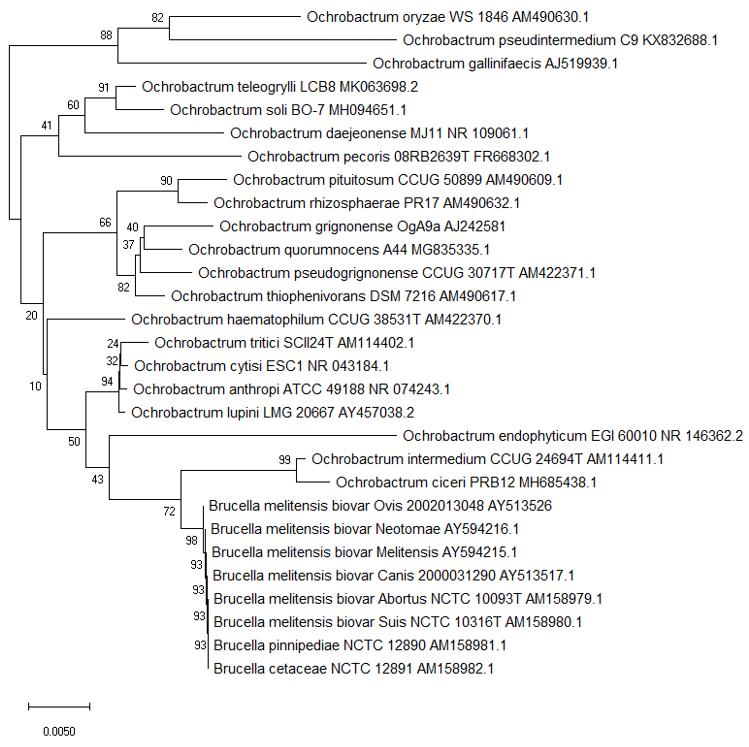
The genus Ochrobactrum emerged from what was previously categorised as the CDC group VD1-2. The type species Ochrobactrum anthropi had previous been called Achromobacter VD based on the Special Bacteriology Section of the US Center for Disease Control [19]. Initial results indicate members of the group grew on MacConkey agar producing catalase, oxidase and urease; strains could be Gram-negative to variable [20]. However, the taxonomic position of Achromobacter became complicated and the name Achromobacter and related CDC group VD were no longer accepted by Bergeys Manual [19] leading to a new classification and the emergence of the genus Ochrobactrum [21]. Ochrobactrum spp. are phylogenetically related to members of the alpha-2 subdivision of Proteobacteria. They are catalogued on the Brucella rRNA branch of rRNA superfamily IV. Thus, from the previous CDC group Vd, a novel genus and a new species, Ochrobactrum anthropi, was proposed [21][22]. The type strain was Gram-negative, aerobic, rod shaped, non-pigmented and motile. It produced acid from a selection of carbohydrates and reduced both nitrate and nitrite and possessed a GC ratio between 56 to 59% [21]. Almost all 56 strains categorised as CDC GroupVD that were used to support the new genus Ochrobactrum came from various human clinical specimens. Since the initial description of O. anthropi, several other species have since been described (Table 1 and Figure 1). Certain Ochrobactrum spp. can be opportunistic pathogens especially in a hospital environment with the majority of reported cases due to hospital-acquired infections in patients with indwelling and invasive medical devices, including central venous catheters and drainage tubes [23]. In addition, the organism shows widespread resistance to penicillins and other antibiotics that cause clinical management issues with immunocompromised hosts [24][25]. The phylogenetic relationship between all described Ochrobactrum spp can be seen in Figure 1.
Table 1. List of current accepted Ochrobactrum species.
|
Species |
Isolation |
Genome Sequences |
Reference |
|
Ochrobactrum anthropi |
Clinical isolate |
Strain: OAB; Size: 4.9 Mbp; Ref Genome: GCA_000742955.1 (41 genomes) |
[22] |
|
Ochrobactrum ciceri |
Nodules of Cicer |
No Genome |
[26] |
|
Ochrobactrum cytisi |
Cystisus nodules |
Strain: IPA7.2; Size: 5.96 Mpb; Ref Genome: GCA_001876955.1 (1 genome) |
[27] |
|
Ochrobactrum daejeonense |
Sludge |
Strain: JCM 16234;; Size: 4.8 Mbp, Ref Genome: GCA_012103095.1 (1 genome) |
[28] |
|
Ochrobactrum endophyticum |
Roots of Glycyrrhiza |
No Genome |
[29] |
|
Ochrobactrum gallinifaecis |
Chicken faeces |
Strain: ISO196; Ref Genome: GCF_006476605.1; Size: 3.74 Mbp (1 genome) |
[11] |
|
Ochrobactrum grignonense |
Wheat Roots |
Strain: OgA9a; Size: 4.84 Mbp; Ref Genome: NZ_NNRL00000000.1 (1 genome) |
[9] |
|
Ochrobactrum haematophilum |
Clinical Isolate |
Strain: LISuc1; Size: 4.91 Mbp; Ref Genome: GCA_003550135.1 (3 genomes) |
[30] |
|
Ochrobactrum intermedium |
Human blood |
Strain: NCTC12171; Size: 4.73 Mbp; Ref Genome: GCA_900454225.1 (18 genomes) |
[22]
|
|
Ochrobactrum lupini |
Lupinus albus rhizosphere |
Strain: LUP21; Size: 5.5 Mbp; Ref Genome: GCA_002252535.1 (2 genomes) |
|
|
Ochrobactrum oryzae |
Rice rhizosphere |
Strain: OA447; Size: 4.47 Mbp; Ref Genome: NZ_PTRC00000000.1 (1 genome) |
[33] |
|
Ochrobactrum pecoris |
Farm Animals |
Strain: 08RB2639; Size: 5.06 Mbp; Ref Genome: GCA_006376675.1 (1 genome) |
[34] |
|
Ochrobactrum pituitosum |
Industrial Environment |
Strain: AA2 Size: 5.47 Mbp; Ref Genome: GCA_002025625.1 (4 genomes) |
[35] |
|
Ochrobactrum pseudintermedium |
Clinical isolate |
Strain: CCUG34735; Size: 4.39 Mbp; Ref Genome: GCA_008932435.1 (1 genome) |
[36] |
|
Ochrobactrum pseudogrignonense |
Clinical isolate |
Strain: K8; Size: 4.99 Mbp; Ref Genome: GCA_001652485.1 (6 genomes) |
[30] |
|
Ochrobactrum quorumnocens |
Potato rhizosphere |
Strain: A44; Size: 5.5 Mbp; Ref Genome: GCA_002278035.1 (2 genomes) |
[37] |
|
Ochrobactrum rhizosphaerae |
Potato rhizosphere |
Strain: PR17; Size: 4.9 Mbp; Ref Genome: GCF_002252475.1 (2 genomes) |
[38] |
|
Ochrobactrum soli |
Cattle farm soil |
Strain: BO-7; Size: 45 Mbp; Ref Genome: GCA_003664555.1 (3 genomes) |
[39] |
|
Ochrobactrum thiophenivorans |
Industrial Environment |
Strain: DSM 7216; Size: 4.4 Mbp; Ref Genome: GCA_002252445.1 (2 genomes) |
[38] |
|
Ochrobactrum teleogrylli |
insect Teleogryllus occipitalis |
No Genome |
[40] |
|
Ochrobactrum tritici |
wheat rhizosphere root soil |
Strain: DSM 13340; Size: 5.5 Mbp; Ref Genome: GCA_012395245.1 (6 genomes) |
[9] |
* First described as Ochrobactrum lupini by Trujillo et al. [31] and later reclassified as Ochrobactrum anthropi by Volpiano et al. [32] following whole-genome sequence analysis.
Figure 1. Phylogenetic structure of the genus Ochrobactrum along with the genus Brucella. (a) The tree based on partial 16S rRNA gene sequences obtained using neighbour joining with Maximum Composite Likelihood method (MEGA package). GenBank accession numbers are given with the species name. Numbers at nodes are bootstrap values based on 1000 resamplings. Bar, 0.0050 substitutions per site [41][42].
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms8111797
References
- Ryan, M.P.; Adley, C.C. Ralstonia: Emerging global opportunistic pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 291–304.
- Ryan, M.P.; Pembroke, J.T.; Adley, C.C. Ralstonia pickettii: A persistent Gram-negative nosocomial infectious organism. J. Hosp. Infect. 2006, 62, 278–284.
- Ryan, M.P.; Adley, C.C. Sphingomonas paucimobilis: A persistent Gram-negative nosocomial infectious organism. Hosp. Infect. 2010, 75, 153–157.
- Coughlan, A.; Ryan, M.P.; Cummins, N.M.; Towler, M.R. The response of Pseudomonas aeruginosa biofilm to the presence of a glass polyalkenoate cement formulated from a silver containing glass. Mater. Sci. 2011, 46, 285–287.
- Ryan, M.P.; Pembroke, J.T. Brevundimonas spp: Emerging global opportunistic pathogens. Virulence 2018, 9, 480–493.
- Jelveh, N.; Cunha, B.A. Ochrobactrum anthropi Hear. Lung J. Acute Crit. Care 1999, 28, 145–146.
- Handschuh, H.; Ryan, M.P.; O’Dwyer, J.; Adley, C.C. Assessment of the bacterial diversity of aircraft water: Identification of the frequent fliers. PLoS ONE 2017, 12, e0170567.
- Möller, L.V.M.; Arends, J.P.; Harmsen, H.J.M.; Talens, A.; Terpstra, P.; Slooff, M.J.H. Ochrobactrum intermedium infection after liver transplantation. Clin. Microbiol. 1999, 37, 241–244.
- Lebuhn, M.; Achouak, W.; Schloter, M.; Berge, O.; Meier, H.; Barakat, M.; Hartmann, A.; Heulin, T. Taxonomic characterization of Ochrobactrum isolates from soil samples and wheat roots, and description of Ochrobactrum tritici sp. nov. and Ochrobactrum grignonense sp. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 2207–2223.
- Goris, J.; Boon, N.; Lebbe, L.; Verstraete, W.; De Vos, P. Diversity of activated sludge bacteria receiving the 3-chloroaniline-degradative plasmid pC1gfp. FEMS Microbiol. Ecol. 2003, 46, 221–230.
- Kämpfer, P.; Buczolits, S.; Albrecht, A.; Busse, H.J.; Stackebrandt, E. Towards a standardized format for the description of a novel species (of an established genus): Ochrobactrum gallinifaecis nov. Int. J. Syst. Evol. Microbiol. 2003, 53, 893–896.
- Bathe, S.; Achouak, W.; Hartmann, A.; Heulin, T.; Schloter, M.; Lebuhn, M. Genetic and phenotypic microdiversity of Ochrobactrum FEMS Microbiol. Ecol. 2006, 56, 272–280.
- El-Sayed, W.S.; Ibrahim, M.K.; Abu-Shady, M.; El-Beih, F.; Ohmura, N.; Saiki, H.; Ando, A. Isolation and Identification of a Novel Strain of the Genus Ochrobactrum with Phenol-Degrading Activity. J. Biosci. Bioeng. 2003, 96, 310–312.
- Sultan, S.; Hasnain, S. Reduction of toxic hexavalent chromium by Ochrobactrum intermedium strain SDCr-5 stimulated by heavy metals. Technol. 2007, 98, 340–344.
- Wu, Y.; He, T.; Zhong, M.; Zhang, Y.; Li, E.; Huang, T.; Hu, Z. Isolation of marine benzo[a]pyrene-degrading Ochrobactrum BAP5 and proteins characterization. J. Environ. Sci. 2009, 21, 1446–1451.
- Zhang, X.H.; Zhang, G.S.; Zhang, Z.H.; Xu, J.H.; Li, S.P. Isolation and characterization of a dichlorvos-degrading strain DDV-1 of Ochrobactrum Pedosphere 2006, 16, 64–71.
- Kettaneh, A.; Weill, F.X.; Poilane, I.; Fain, O.; Thomas, M.; Herrmann, J.L.; Hocqueloux, L. Septic shock caused by Ochrobactrum anthropi in an otherwise healthy host. Clin. Microbiol. 2003, 41, 1339–1341.
- Ozdemir, D.; Soypacaci, Z.; Sahin, I.; Bicik, Z.; Sencan, I. Ochrobactrum anthropi endocarditis and septic shock in a patient with no prosthetic valve or rheumatic heart disease: Case report and review of the literature. J. Infect. Dis. 2006, 59, 264–265.
- Chester, B.; Cooper, L.H. Achromobacter species (CDC group Vd): Morphological and biochemical characterization. Clin. Microbiol. 1979, 9, 425–436.
- Barson, W.J.; Cromer, B.A.; Marcon, M.J. Puncture wound osteochondritis of the foot caused by CDC group Vd. Clin. Microbiol. 1987, 25, 2014–2016.
- Holmes, B.; Popoff, M.; Kiredjian, M.; Kersters, K. Ochrobactrum anthropi nov., sp. nov. from human clinical specimens and previously known as group Vd. Int. J. Syst. Bacteriol. 1988, 38, 406–416.
- Velasco, J.; Romero, C.; López-Goñi, I.; Leiva, J.; Dĩaz, R.; Moriyón, I. Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium nov., a new species with a closer relationship to Brucella spp. Int. J. Syst. Bacteriol. 1998, 48, 759–768.
- Rastogi, N.; Mathur, P. Ochrobactrum anthropi: An emerging pathogen causing meningitis with sepsis in a neurotrauma patient. Infect. Dev. Ctries. 2017, 11, 733–735.
- Nadjar, D.; Labia, R.; Cerceau, C.; Bizet, C.; Philippon, A.; Arlet, G. Molecular characterization of chromosomal class C β-lactamase and its regulatory gene in Ochrobactrum anthropi. Agents Chemother. 2001, 45, 2324–2330.
- Soloaga, R.; Carrion, N.; Pidone, J.; Guelfand, L.; Margari, A.; Altieri, R. Bacteremia relacionada a cateter por Ochrobactrum anthropi. Medicina (Buenos Aires) 2009, 69, 655–657.
- Imran, A.; Hafeez, F.Y.; Frühling, A.; Schumann, P.; Malik, K.A.; Stackebrandt, E. Ochrobactrum ciceri nov., isolated from nodules of Cicer arietinum. Int. J. Syst. Evol. Microbiol. 2010, 60, 1548–1553.
- Zurdo-Piñeiro, J.L.; Rivas, R.; Trujillo, M.E.; Vizcaíno, N.; Carrasco, J.A.; Chamber, M.; Palomares, A.; Mateos, P.F.; Martínez-Molina, E.; Velázquez, E. Ochrobactrum cytisi nov., isolated from nodules of Cytisus scoparius in Spain. Int. J. Syst. Evol. Microbiol. 2007, 57, 784–788.
- Woo, S.G.; Ten, L.N.; Park, J.; Lee, M. Ochrobactrum daejeonense nov., a nitrate-reducing bacterium isolated from sludge of a leachate treatment plant. Int. J. Syst. Evol. Microbiol. 2011, 61, 690–2696.
- Li, L.; Li, Y.Q.; Jiang, Z.; Gao, R.; Nimaichand, S.; Duan, Y.Q.; Egamberdieva, D.; Chen, W.; Li, W.J. Ochrobactrum endophyticum nov., isolated from roots of Glycyrrhiza uralensis. Arch. Microbiol. 2016, 198, 171–179.
- Kämpfer, P.; Scholz, H.C.; Huber, B.; Falsen, E.; Busse, H.J. Ochobactrum haematophilum nov. and Ochrobactrum pseudogrignonense sp. nov., isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 2007, 57, 2513–2518.
- Trujillo, M.E.; Willems, A.; Abril, A.; Planchuelo, A.M.; Rivas, R.; Ludeña, D.; Mateos, P.F.; Martínez-Molina, E.; Velázquez, E. Nodulation of Lupinus albus by Strains of Ochrobactrum lupini nov. Appl. Environ. Microbiol. 2005, 71, 1318–1327.
- Volpiano, C.G.; Sant’anna, F.H.; Ambrosini, A.; Lisboa, B.B.; Vargas, L.K.; Passaglia, L.M.P. Reclassification of Ochrobactrum lupini as a later heterotypic synonym of Ochrobactrum anthropi based on whole-genome sequence analysis. J. Syst. Evol. Microbiol. 2019, 69, 2312–2314.
- Tripathi, A.K.; Verma, S.C.; Chowdhury, S.P.; Lebuhn, M.; Gattinger, A.; Schloter, M. Ochrobactrum oryzae nov., an endophytic bacterial species isolated from deep-water rice in India. Int. J. Syst. Evol. Microbiol. 2006, 56, 1677–1680.
- Kämpfer, P.; Huber, B.; Busse, H.J.; Scholz, H.C.; Tomaso, H.; Hotzel, H.; Melzer, F. Ochrobactrum pecoris nov., isolated from farm animals. Int. J. Syst. Evol. Microbiol. 2011, 61, 2278–2283.
- Huber, B.; Scholz, H.C.; Kämpfer, P.; Falsen, E.; Langer, S.; Busse, H.J. Ochrobactrum pituitosum nov., isolated from an industrial environment. Int. J. Syst. Evol. Microbiol. 2010, 60, 321–326.
- Teyssier, C.; Marchandin, H.; Jean-Pierre, H.; Masnou, A.; Dusart, G.; Jumas-Bilak, E. Ochrobactrum pseudintermedium nov., a novel member of the family Brucellaceae, isolated from human clinical samples. Int. J. Syst. Evol. Microbiol. 2007, 57, 1007–1013.
- Krzyżanowska, D.M.; Maciąg, T.; Ossowicki, A.; Rajewska, M.; Kaczyński, Z.; Czerwicka, M.; Rąbalski, Ł.; Czaplewska, P.; Jafra, S. Ochrobactrum quorumnocens Nov., a quorum quenching bacterium from the potato rhizosphere, and comparative genome analysis with related type strains. PLoS ONE 2019, 14, e0210874.
- Kämpfer, P.; Sessitsch, A.; Schloter, M.; Huber, B.; Busse, H.J.; Scholz, H.C. Ochrobactrum rhizosphaerae nov. and Ochrobactrum thiophenivorans sp. nov., isolated from the environment. Int. J. Syst. Evol. Microbiol. 2008, 58, 1426–1431.
- Choi, G.M.; Kim, K.M.; Yun, C.S.; Lee, S.Y.; Kim, S.Y.; Wee, J.H.; Im, W.T. Ochrobactrum soli nov., Isolated from a Korean cattle farm. Curr. Microbiol. 2020, 77, 1104–1110.
- Hu, M.; Li, X.; Li, Z.; Liu, B.; Yang, Z.; Tian, Y. Ochrobactrum teleogrylli nov., a pesticide-degrading bacterium isolated from the insect Teleogryllus occipitalis living in deserted cropland. Int. J. Syst. Evol. Microbiol. 2020, 70, 2217–2225.
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Biol. Evol. 2018, 35, 1547–1549.
- Ryan, M.P.; Adley, C.C.; Pembroke, J.T. The use of MEGA as an educational tool for examining the phylogeny of antibiotic resistance genes. In Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education,; Méndez-Vilas, A., Ed.; Formatex: Badajoz, Spain, 2013; Volume 4 pp. 736–743.