Neurodegenerative disorders and cancer may appear unrelated illnesses. Yet, epidemiologic studies indicate an inverse correlation between their respective incidences for specific cancers. Possibly explaining these findings, increasing evidence indicates that common molecular pathways are involved, often in opposite manner, in the pathogenesis of both disease families. Genetic mutations in the MAPT gene encoding for TAU protein cause an inherited form of frontotemporal dementia, a neurodegenerative disorder, but also increase the risk of developing cancer. Assigning TAU at the interface between cancer and neurodegenerative disorders, two major aging-linked disease families, offers a possible clue for the epidemiological observation inversely correlating these human illnesses. In addition, the expression level of TAU is recognized as a prognostic marker for cancer, as well as a modifier of cancer resistance to chemotherapy. Because of its microtubule-binding properties, TAU may interfere with the mechanism of action of taxanes, a class of chemotherapeutic drugs designed to stabilize the microtubule network and impair cell division. Indeed, a low TAU expression is associated to a better response to taxanes. Although TAU main binding partners are microtubules, TAU is able to relocate to subcellular sites devoid of microtubules and is also able to bind to cancer-linked proteins, suggesting a role of TAU in modulating microtubule-independent cellular pathways associated to oncogenesis. This concept is strengthened by experimental evidence linking TAU to P53 signaling, DNA stability and protection, processes that protect against cancer.
- neurodegeneration
- tauopathies
- cancer
- TAU protein
- DNA protection
1. Coming Together: Cancer and Neurodegenerative Disorders, Do They Share Dysregulated Pathways?
The fundamental defect resulting in cancer is an aberrant molecular machinery controlling cell division and cell death. Rather than responding appropriately to the signals that restrain cell growth, neoplastic cells divide and invade normal tissues with the potential to colonize multiple organs. In contrast, differentiated neurons display specific molecular and morphological signatures that prevent them from further cell division. However, post-mitotic neurons respond to stress conditions such as trophic factor deprivation, oxidative overload or DNA damage by up-regulating cell cycle activators, possibly causing neuronal death [1]. In fact, hallmarks of DNA replication and active cell cycle are observed in post-mitotic neurons of patients suffering of a neurodegenerative process such as in tauopathies [2][3]. This evidence conveys the postulation that neurodegeneration and cancer, despite appearing unrelated human illnesses, may both result from inappropriately regulated cellular pathways, such as cell-cycle control or cell death because of DNA damage [4][5][6].
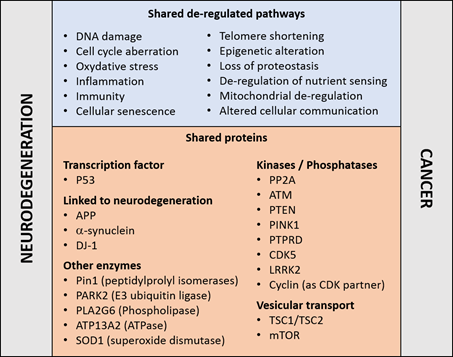
Reinforcing this notion, an established risk factor for cancer and neurodegenerative disorders is aging—a manifestation of a time-dependent accumulation of harmful insults [7]. The two disease families share cellular and molecular hallmarks of aging [8]: genomic instability, DNA damage, epigenetic modifications, nutrient sensing abnormalities, proteostasis unbalance, mitochondrial dysfunction, telomere shortening, cellular senescence, and altered intercellular communication (Figure 1). Moreover, the aberrant regulation of common proteins and cellular pathways may occur in opposite directions. For example, whilst the regulatory mechanisms associated to the tumor suppressor P53 are frequently down-regulated in cancers [9], P53 is upregulated in concomitance to the neurodegenerative process [10][11][12]; and the reverse is true for the tumor promoting protein peptilprolyl isomerase PIN1 [13]. Both examples are discussed in more details below. These observations reinforce the concept that the occurrence of neurodegeneration and cancer may result from the deregulation of genetic factors or proteins implicated in cellular pathways common to both disease families.
Figure 1. Molecular pathways (top) and proteins (bottom) that have been linked to cancer and neurodegenerative disorders, details and references are given in the main text.
Intriguing epidemiological interrelations indicate an inverse association between neurodegenerative disorders and a variety of cancer types, suggesting that a propensity for one family of diseases may decrease the risk for the other. Indeed, cancer survivors present decreased incidence for Alzheimer’s disease (AD), Parkinson’s disease (PD) and Huntington’s disease (HD), and vice versa [14][15][16][17][18][19][20][21][22]. A history of smoking related cancers has a protective impact against AD [14], whereas AD patients are less prone to develop lung cancer [23][24][25][26]. For amyotrophic lateral sclerosis (ALS) a decreased frequency of cancer is observed after disease onset [27], although a cancer diagnosis does not affect the occurrence of ALS [28][29]. In contrast, a positive correlation is observed between cancer and aging-related disorders as stroke, macular degeneration, non-neurodegenerative dementia, and osteoarthritis [30]. This is also true for the positive association of PD with melanoma and prostate cancer [30][31][32][33][34]. Cancer chemotherapies are also associated with a lower incidence of AD [35], and some of them disturb white matter structures and neuronal connectivity [36].
The interpretation of epidemiologic studies is complex and confronted with the challenge of identifying the molecular mechanisms influencing occurrence, pharmacological treatment and ultimately the survival of patients affected by one or the other of the two disorder families [37][38]. Of help is the identification of mutations in genes implicated in both disorders, such as those involved in regulation of cell cycle, DNA repair, oxidative stress, cell death and autophagy [39][40][41][42][43]. In this context, the protein kinase ataxia-telangiectasia mutated (ATM) and PARK2 are two examples. Germinal homozygotes mutations in ATM, a kinase tightly involved in the DNA damage response, cause ataxia-telangiectasia, a neurodegenerative disorder with a high predisposition to cancer [44]. Somatic mutations and deletions of PARK2, an E3-ubiquitin ligase involved in degradation of several target proteins including the cell cycle modulator cyclin E, have been reported in different tumor types [44], whereas germinal mutations in PARK2 are linked to PD. Large genome-wide association studies searching for co-heritability confirm shared genetic risks between AD and cancer with the largest overlap for gene sets annotated as expression regulators [45]. Interestingly, genetic components modulate the risk in the same direction and other in the opposite manner for the two disorders, but, unfortunately, it was not possible to identify in this study single nucleotide polymorphisms due to the involvement of multiple loci. Transcriptomic comparison of three cancer types and three central nervous system disorders further indicates expression deregulation in opposite directions [46]. Incidentally, genes that are strongly associated to neurodegenerative disorders, i.e., because their products are the main constituents of hallmark brain deposits and they may lead to early-onset inherited disease forms, do not exhibit typical features of oncogenes or tumor suppressors but appears to be involved in some processes associated to cancers. For example, APP promotes migration and invasion of breast cancer cells [47] and is a predictor of poor prognosis in some breast cancers [48]; whereas alpha-synuclein may be implicated in the malignant progression of meningioma [49]. A recent analysis of cancer incidence in carriers of FTDP-17 MAPT mutations showed increased risk of developing cancer [50]. The tumor types occurring in FTDP-17 families were variable (hematological, lung, breast, and colorectal cancers) suggesting that mutations in TAU, the protein encoded by the MAPT gene, may present predisposing oncogenic elements for genomic instability without tissue specificity [50]. In agreement with these data is the increased chromosomal aberration detected in lymphocytes and fibroblasts isolated from carriers of FTDP-17 MAPT mutations [51]. Overall, it appears that MAPT mutations are driving factors for neurodegenerative disorders as well as some cancer forms.
2. The TAU Protein
TAU is generally described as a protein highly expressed in the central nervous system. The human brain expresses at least six TAU isoforms with molecular weights ranging from 45 to 65 kDa [52][53] generated by alternative splicing of exons 2, 3 and 10 out of the 16 exons composing the MAPT gene [52]. The number and relative amount of the TAU splice variants vary in a cell type specific manner, during development and depending on the clinical features of neurodegenerative disorders [54][55]. Adding complexity, TAU proteins are modified by a considerable number and variety of posttranslational modifications; which become markedly increased in disease, as e.g., for the hyper-phosphorylated forms characterizing tauopathies [56][57]. Yet, TAU is also present in skeletal muscle, breast, kidney, prostate and in cultured fibroblasts [58][59][60][61][62][63][64][65], and at a lower level in the intestine, skin, liver, and submandibular gland [66]. A detailed analysis of the TAU species present in peripheral tissues was initially performed in rodents [67]. This led to the identification of an additional TAU isoform with a molecular weight >100 kDa (“big TAU”), generated by an unspliced 4a exon, present in rat peripheral tissues and in nearly all central neurons projecting to the periphery [68]. Similar findings were reported in humans [69]. A detailed analysis of TAU expression at the level of mRNA, protein and post-translational modifications is crucial to better demonstrate and understand the role played by TAU in neoplastic disorders.
TAU binds to microtubules and regulates their dynamics, e.g., for the structural organization of axons and the exchange of proteins and cellular organelles between cell soma and the synapse, or for influencing the mitotic spindle. These functions are possible because TAU is a scaffold protein linking a variety of molecular partners under the control of a complex pattern of post-translational protein modifications. A simplistic concept for the role of TAU in neurodegenerative diseases is that its aberrant translational and post-translational modifications cause microtubule dissociation, followed by an increase in the soluble pool driving a toxic gain-of-function characterized by the acquisition of pathogenic conformations, self-assembly, fibril formation, and NFT deposition. This cascade of events is associated with synaptic loss, neuronal dysfunction and cell death. However, soluble TAU may relocate to other subcellular sites. In the neuronal dendrites, TAU has been shown to regulate synaptic plasticity by binding to the proto-oncogene tyrosine-protein kinase FYN, a protein involved in oncogenesis [70] . TAU is also located in the cell nucleus [71][72] and can bind DNA, acquiring DNA protecting properties [73][74] and contributing to regulate chromatin compaction [75]. Additional involvements of nuclear TAU in RNA transcription, retrotransposon mobility, and structural organization of the nucleolus and the nuclear membrane are reported [76][77][78][79]. We recently reported a modulatory effect of TAU on the tumor suppressor P53 and down-stream function such as apoptosis and senescence [80]. All these data support a role of TAU that may be independent to its binding to microtubules and may contribute to cancer. However, TAU is found predominantly bound to microtubules, and also in this function the likely contribution to cancer are well documented.
3. TAU and Microtubule-Targeting Chemotherapy
The mitotic spindle is the critical structure organizing the microtubule scaffold enabling chromosomal segregation and cell division. So, targeting microtubules represent a successful mode of action for cancer chemotherapy. A classic example of this class of drugs are taxanes, which bind beta-tubulin at the microtubule inner surface and inhibit microtubule depolymerization. Through the alteration of the dynamic assembly and disassembly of microtubules, taxanes restrict spindle activity and impair the cell cycle in the G1/G2 phase of mitosis. The cytostatic effect of taxanes results in the subsequent induction of apoptosis, which is partly regulated by the tumor suppressor P53 [81]. The taxane Paclitaxel present in the bark of the Pacific yew tree, is produced in a semisynthetic way from Taxus baccata, and is used in clinical oncology since almost three decades [82]. The resistance to taxanes observed in certain cancer types frequently limits the therapeutic efficacy. Possible causes include the action of xenobiotic efflux pumps, alterations in apoptotic and signal transduction pathways, and abnormalities in target engagement modulated by microtubule interacting proteins [83]. The microtubule-binding protein TAU may interfere with the binding of taxanes to tubulin [84]. Consequently, increased cellular concentration of TAU or its affinity to microtubules are considered factors protecting microtubules against taxane therapy [85][86][87], and are thus assessed as predictors of therapeutic efficacy for microtubule-targeting drugs [88]. For example, MAPT is the most differentially expressed gene as a function of response to preoperative Paclitaxel treatment in breast cancer , whereby low TAU mRNA predicted complete response to taxanes, as confirmed also in additional studies [89]. In estrogen receptor (ER)-negative breast cancer, the correlation between low TAU expression and ER status may explain the higher sensitivity to Paclitaxel. Low TAU reflected by a better response to taxanes is reported also in ovarian [90][91], gastric [92], prostate [93] and non-small-cell lung cancer [94]. Notably, retinoic acid-induced TAU expression in neuroblastoma cells results in increased resistance to Paclitaxel [95], although this may be related to their differentiation state. These results feed the concept that anti-TAU drugs may be exploited as a strategy to improve the outcome of taxane-based chemotherapies. Nevertheless, some studies came to an opposite conclusion and some Paclitaxel trials did not confirm the predictive value of TAU determination [96][97][98]. The discordance between these studies may result from the choice of chemotherapy regimen, the taxane used, the cancer type, and possibly from the limitation imposed by the analysis of a single marker. Additional insights were gained by employing cellular models. Taxane-resistant prostate cells express higher level of TAU compared to parental lines, whereby TAU modulation of PI3K signaling may play a role [99]. The microRNA miR-34c-5p regulates MAPT gene expression in gastric cancer cell lines thereby modulating the sensitivity to Paclitaxel [100], whereas in non-small cell lung cancer cells the same effect was modulated by miR-186 [101]. The selective ER inhibitor Fulvestrant, in contrast to Tamoxifen, reduces all TAU protein isoforms and increases taxane sensitivity in ER-positive breast cancer cells. It is concluded that modulation of TAU expression impacts the response to taxanes in cancer cells from diverse origins. An example that qualified TAU as a potential therapeutic agent is indeed based on its microtubule-binding modulation of the mitotic spindle. The use of a tailored protein fusion between epidermal growth factor (EGF; targeting component) and TAU (effector component) resulted in a cytostatic and apoptotic response in epidermal growth factor receptor (EGFR)-positive pancreatic cancer cells [102], a finding confirmed in other models [103].
4. TAU as a Prognostic Marker in Cancer
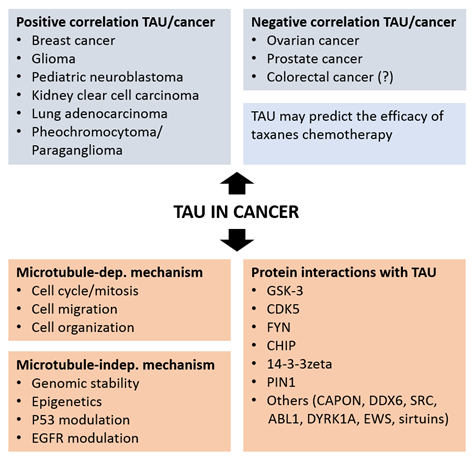
The analysis of MAPT gene transcription and TAU protein expression in healthy and neoplastic tissues supports a role of TAU in cancer. This analytical work, in part performed in silico on available cancer databases, defines a value for TAU as a prognostic marker in various cancers (Figure 2). The following paragraphs review the outcome of these studies for distinct cancer types.
Figure 2. Positive and negative association of TAU expression with different type of cancers (top panels) and mechanisms and protein interactions associating TAU to cancer.
In breast cancer, higher TAU protein expression is associated to a better outcome and survival independently to the therapy [104]. However, TAU level did not correlate with tumor size or nodal status or patient age. A positive correlation between TAU expression and the receptors for estrogen and progesterone (PR) expression was confirmed in multiple studies, in particular for low grade, ER/PR-positive, and human epidermal growth factor receptor 2 (HER2)-negative cancers [104][105][106]. An inducible imperfect estrogen response element was identified upstream of the MAPT promoter [106][107][108][109][110][111][112], which is consistent with the endocrine sensitivity of TAU- and ER-positive tumors. Among a panel of breast cancer cell lines with different levels of TAU mRNA and TAU isoforms, down-regulation of ER expression and the presence of ER inhibitors affected TAU expression in a cell-specific manner [113][114]. The inverse correlation TAU/HER2 is remarkable due to the proximity of the two genes in the 17q12 chromosomic region. A thorough analysis of the cancer genome atlas (TCGA) cohorts in tumors with high or low TAU expression, demonstrates a positive correlation between MAPT transcription and overall survival of patients with breast cancer [115]. However, a study aiming at understanding how circulating tumor cells reattach in distant tissue indicate that in metastatic breast tumor TAU is more expressed and that TAU microtubule binding is necessary and sufficient to promote tumor cell reattachment [116].
For ovarian cancer, immune histochemical analysis shows that the three-year survival was significantly higher in the TAU-negative when compared to the TAU-positive group. These data suggest, in contrast to breast cancer, that high TAU expression is associated with an unfavorable prognostic. However, the results were not confirmed in the TCGA cohorts [115], which is based on gene transcript assessment rather than on protein determination. In view of the complex regulation of TAU protein homeostasis at the level of translation and post-translational modification, a careful TAU protein analysis may be more informative in this context. Notably, the endometrioid carcinoma TOV112D cells showed the highest TAU protein expression among a panel of ovarian cancer cell lines and TAU knock-down inhibited cell proliferation, in accordance with the favorable prognostic associated to low TAU expression.
An early study in prostate cancer found that TAU protein overexpression was associated with a better prognostic (lower Gleason score) in a cohort of 30 patients [117]. The use of a dephosphorylated-specific TAU antibody, demonstrated the absence of phosphorylation at the Tau-1 epitope in neoplastic prostate tissue [117]. Immune histochemical analysis on a tissue microarray containing 17,747 prostate samples showed under the selected experimental conditions detectable TAU expression in 8% of the cancer samples and no measurable TAU in the normal tissue, evidence for TAU overexpression as a moderate prognostic feature in a small prostate cancer subset [118]. TAU expression was associated with advanced tumor stage, high Gleason score, positive nodal stage, and risk for recurrence in all cancers independently of the erythroblast transformation specific-related gene (ERG) status [118]. About half of prostate cancers are due to gene fusions linking the androgen-regulated transmembrane protease TMPRSS2 with the transcription factor ERG [119][120] resulting in a massive androgen-dependent overexpression of ERG. Other somatic mutations associated to prostate cancer include PTEN genomic deletions, which positively associate to TAU expression with the highest MAPT transcription observed in ERG positive cancers. This observation is possibly linked to the suggested regulatory function in microtubule dynamics of ERG [121][122], which binds and stabilizes soluble tubulin [123]. The association between high TAU expression and poor overall survival was confirmed in an independent study [124] also describing an inverse interaction between MAPT and PTEN in prostate cancer. However, the transcriptomic-based TCGA cohorts failed to show a positive or negative association between TAU expression and survival in the prostate cancer cohort. A detailed analysis of TAU in prostate cancer cell lines, revealed high expression of multiple TAU splice variants, including big TAU and a previously undescribed variant, in comparison to e.g., the primarily fetal TAU isoform present in human neuroblastoma SH-SY5Y cells [125][126] or the six main isoforms described in normal adult human brain. Moreover, the TAU phosphorylation pattern observed in prostate cancer cells reflects what observed in tauopathies when compared to healthy adult brain with a large proportion of TAU not bound to microtubules. Association of TAU to phosphoinositide 3 kinase (PI3K) suggests a microtubule-independent mechanism possibly linked to cell signaling [127]. Consistent with this, in docetaxel-resistant prostate cell lines [128] TAU down-regulation inhibits cell proliferation by the PI3K/mTOR signaling pathway.
Analysis of the bottom and top 20% MAPT expressers in pediatric neuroblastoma revealed a better prognosis for the top quintile according to the MAPT transcript analyzed on microarray (NCBO BioPortal) [129]. The data were substantiated with a significant correlation with apoptotic-and proliferation-linked genes. In contrast, increased survival was not associated to the mRNA for alpha-synuclein, another neurodegeneration-associated protein [129].
The value of TAU as a biomarker for disease-free survival rate in glioma (TCGA data set) was shown by comparing the bottom and top 20% MAPT transcript expressers [130]. Moreover, the histological tumor grade was inversely correlated with TAU expression. Consistent with these data, in the TAU mRNA-top quintile group, transcriptional activity was higher for pro-apoptotic genes and lower for proliferation-associated genes. Evidence that transcription alterations for genes associated with neurodegeneration—with the exception of MAPT—are not common drivers of gliomas was confirmed in another study, suggesting an important role of TAU in slowing down or preventing the clinical evolution of these tumors [131]. Histochemical analysis showed that cells from low malignancy glioma display increase TAU protein expression, with the inverse observation for cells from more aggressive tumors.
In colorectal cancer, CpG island hypermethylation in MAPT is found in about a quarter of the samples in a cohort with hundred stage II patients, but it was absent in normal colorectal mucosa [132]. This study was inspired by the presence of methylation in the MAPT promoter in AD [133], PD [134] as well as prostate cancer [135]. MAPT hypermethylation is a marker for lower five-year survival indicating that, similarly to breast cancer, low TAU expression is linked to a worse prognostic in both cancers. However, analysis of the TCGA database did not confirm the data . At the protein level, increased TAU phosphorylation at Ser199/202 is a predictor of non-metastatic colon cancer [136]. Consistent with a main hypothesis for AD, hyperphosphorylated forms of TAU with impaired microtubule binding were reported in colorectal cell lines [137].
TAU appears implicated in Bloom’s syndrome, a rare genetic disorder resulting from homozygous mutations of the BLM gene with a high rate of spontaneous chromosome abnormalities and predisposition to cancer [138]. Mutated BLM cells experience replication stress and display chromosome segregation defects, but continue to divide indicating a tolerance for DNA damage. TAU was identified in a genome-wide RNAi screen and transcriptomic analysis as a critical protein enabling this phenotype. Indeed, TAU overexpressing Bloom’s syndrome cells undergo cell death when TAU is down-regulated [138]. This is interpreted as TAU acting as a negative regulator of DNA damage-induced cell death.
A comprehensive analysis of the TCGA cohorts shows positive association between TAU mRNA expression and survival also in kidney clear cell carcinoma, lung adenocarcinoma, pheochromocytoma/paraganglioma. In contrast, a negative association is found for colon and head and neck cancers.
The clinical and prognostic value of TAU analyzed at the mRNA and protein level has been investigated for many tumors with results crucially dependent on the cancer type (Figure 2). Whether the correlative studies implicating TAU in cancer will eventually demonstrate an active participation of TAU in oncogenesis requires undoubtedly further experimental evidence. As of today, the mechanisms that may explain if and how TAU differentially impact tumor cell aggressiveness in different cancer types remains at large poorly understood. As commented previously, transcriptome analysis does not take into account the pathogenic effects of protein homeostasis, which in the case of TAU is complex and tightly associated to disease. As in the case of neurodegenerative tauopathies, a detailed characterization of MAPT transcription and translation as well as the biochemical characterization of TAU protein including its modification, cellular distribution and interacting proteins, is now necessary in the studies linking TAU to cancer.
This entry is adapted from the peer-reviewed paper 10.3390/brainsci10110862
References
- Currais, A.; Goldberg, J.; Farrokhi, C.; Chang, M.; Prior, M.; Dargusch, R.; Daugherty, D.; Armando, A.; Quehenberger, O.; Maher, P.; et al. A comprehensive multiomics approach toward understanding the relationship between aging and dementia. Aging (Albany NY) 2015, 7, 937–955, doi:10.18632/aging.100838.
- Bonda, D.J.; Lee, H.P.; Kudo, W.; Zhu, X.; Smith, M.A.; Lee, H.G. Pathological implications of cell cycle re-entry in Alzheimer disease. Expert Rev. Mol. Med. 2010, 12, e19, doi:10.1017/s146239941000150x.
- McShea, A.; Lee, H.G.; Petersen, R.B.; Casadesus, G.; Vincent, I.; Linford, N.J.; Funk, J.O.; Shapiro, R.A.; Smith, M.A. Neuronal cell cycle re-entry mediates Alzheimer disease-type changes. Biochim. Biophys. Acta 2007, 1772, 467–472, doi:10.1016/j.bbadis.2006.09.010.
- Kennedy, S.R.; Loeb, L.A.; Herr, A.J. Somatic mutations in aging, cancer and neurodegeneration. Mech. Ageing Dev. 2012, 133, 118–126, doi:10.1016/j.mad.2011.10.009.
- Lovell, M.A.; Markesbery, W.R. Oxidative DNA damage in mild cognitive impairment and late-stage Alzheimer’s disease. Nucleic Acids Res. 2007, 35, 7497–7504, doi:10.1093/nar/gkm821.
- Mullaart, E.; Boerrigter, M.E.; Ravid, R.; Swaab, D.F.; Vijg, J. Increased levels of DNA breaks in cerebral cortex of Alzheimer’s disease patients. Neurobiol. Aging 1990, 11, 169–173, doi:10.1016/0197-4580(90)90542-8.
- Moskalev, A.A.; Shaposhnikov, M.V.; Plyusnina, E.N.; Zhavoronkov, A.; Budovsky, A.; Yanai, H.; Fraifeld, V.E. The role of DNA damage and repair in aging through the prism of Koch-like criteria. Ageing Res. Rev. 2013, 12, 661–684, doi:10.1016/j.arr.2012.02.001.
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217, doi:10.1016/j.cell.2013.05.039.
- Meek, D.W. Regulation of the p53 response and its relationship to cancer. Biochem. J. 2015, 469, 325–346, doi:10.1042/bj20150517.
- Bae, B.I.; Xu, H.; Igarashi, S.; Fujimuro, M.; Agrawal, N.; Taya, Y.; Hayward, S.D.; Moran, T.H.; Montell, C.; Ross, C.A.; et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington’s disease. Neuron 2005, 47, 29–41, doi:10.1016/j.neuron.2005.06.005.
- Bretaud, S.; Allen, C.; Ingham, P.W.; Bandmann, O. p53-dependent neuronal cell death in a DJ-1-deficient zebrafish model of Parkinson’s disease. J. Neurochem. 2007, 100, 1626–1635, doi:10.1111/j.1471-4159.2006.04291.x.
- Hooper, C.; Meimaridou, E.; Tavassoli, M.; Melino, G.; Lovestone, S.; Killick, R. p53 is upregulated in Alzheimer’s disease and induces tau phosphorylation in HEK293a cells. Neurosci. Lett. 2007, 418, 34–37, doi:10.1016/j.neulet.2007.03.026.
- Driver, J.A.; Zhou, X.Z.; Lu, K.P. Pin1 dysregulation helps to explain the inverse association between cancer and Alzheimer’s disease. Biochim. Biophys. Acta 2015, 1850, 2069–2076, doi:10.1016/j.bbagen.2014.12.025.
- Driver, J.A.; Beiser, A.; Au, R.; Kreger, B.E.; Splansky, G.L.; Kurth, T.; Kiel, D.P.; Lu, K.P.; Seshadri, S.; Wolf, P.A. Inverse association between cancer and Alzheimer’s disease: Results from the Framingham Heart Study. BMJ 2012, 344, e1442, doi:10.1136/bmj.e1442.
- Driver, J.A.; Logroscino, G.; Buring, J.E.; Gaziano, J.M.; Kurth, T. A prospective cohort study of cancer incidence following the diagnosis of Parkinson’s disease. Cancer Epidemiol. Biomark. 2007, 16, 1260–1265, doi:10.1158/1055-9965.epi-07-0038.
- Inzelberg, R.; Jankovic, J. Are Parkinson disease patients protected from some but not all cancers? Neurology 2007, 69, 1542–1550, doi:10.1212/01.wnl.0000277638.63767.b8.
- Møller, H.; Mellemkjaer, L.; McLaughlin, J.K.; Olsen, J.H. Occurrence of different cancers in patients with Parkinson’s disease. BMJ 1995, 310, 1500–1501, doi:10.1136/bmj.310.6993.1500.
- Roe, C.M.; Fitzpatrick, A.L.; Xiong, C.; Sieh, W.; Kuller, L.; Miller, J.P.; Williams, M.M.; Kopan, R.; Behrens, M.I.; Morris, J.C. Cancer linked to Alzheimer disease but not vascular dementia. Neurology 2010, 74, 106–112, doi:10.1212/WNL.0b013e3181c91873.
- Shi, H.B.; Tang, B.; Liu, Y.W.; Wang, X.F.; Chen, G.J. Alzheimer disease and cancer risk: A meta-analysis. J. Cancer Res. Clin. Oncol. 2015, 141, 485–494, doi:10.1007/s00432-014-1773-5.
- Sørensen, S.A.; Fenger, K.; Olsen, J.H. Significantly lower incidence of cancer among patients with Huntington disease: An apoptotic effect of an expanded polyglutamine tract? Cancer 1999, 86, 1342–1346.
- Tabarés-Seisdedos, R.; Dumont, N.; Baudot, A.; Valderas, J.M.; Climent, J.; Valencia, A.; Crespo-Facorro, B.; Vieta, E.; Gómez-Beneyto, M.; Martínez, S.; et al. No paradox, no progress: Inverse cancer comorbidity in people with other complex diseases. Lancet Oncol. 2011, 12, 604–608, doi:10.1016/s1470-2045(11)70041-9.
- Majd, S.; Power, J.; Majd, Z. Alzheimer’s Disease and Cancer: When Two Monsters Cannot Be Together. Front. Neurosci. 2019, 13, doi:10.3389/fnins.2019.00155.
- Greco, A.; Sanchez Valle, J.; Pancaldi, V.; Baudot, A.; Barillot, E.; Caselle, M.; Valencia, A.; Zinovyev, A.; Cantini, L. Molecular Inverse Comorbidity between Alzheimer’s Disease and Lung Cancer: New Insights from Matrix Factorization. Int. J. Mol. Sci. 2019, 20, 3114, doi:10.3390/ijms20133114.
- Musicco, M.; Adorni, F.; Di Santo, S.; Prinelli, F.; Pettenati, C.; Caltagirone, C.; Palmer, K.; Russo, A. Inverse occurrence of cancer and Alzheimer disease: A population-based incidence study. Neurology 2013, 81, 322–328, doi:10.1212/WNL.0b013e31829c5ec1.
- Ou, S.M.; Lee, Y.J.; Hu, Y.W.; Liu, C.J.; Chen, T.J.; Fuh, J.L.; Wang, S.J. Does Alzheimer’s Disease Protect against Cancers? A Nationwide Population-Based Study. Neuroepidemiology 2013, 40, 42–49, doi:10.1159/000341411.
- Sánchez-Valle, J.; Tejero, H.; Ibáñez, K.; Portero, J.L.; Krallinger, M.; Al-Shahrour, F.; Tabarés-Seisdedos, R.; Baudot, A.; Valencia, A. A molecular hypothesis to explain direct and inverse co-morbidities between Alzheimer’s Disease, Glioblastoma and Lung cancer. Sci. Rep. 2017, 7, 4474, doi:10.1038/s41598-017-04400-6.
- Gibson, S.B.; Abbott, D.; Farnham, J.M.; Thai, K.K.; McLean, H.; Figueroa, K.P.; Bromberg, M.B.; Pulst, S.M.; Cannon-Albright, L. Population-based risks for cancer in patients with ALS. Neurology 2016, 87, 289–294, doi:10.1212/WNL.0000000000002757.
- Fang, F.; Al-Chalabi, A.; Ronnevi, L.O.; Turner, M.R.; Wirdefeldt, K.; Kamel, F.; Ye, W. Amyotrophic lateral sclerosis and cancer: A register-based study in Sweden. Amyotroph. Lateral Scler. Front. Degener. 2013, 14, 362–368, doi:10.3109/21678421.2013.775309.
- Freedman, D.M.; Wu, J.; Daugherty, S.E.; Kuncl, R.W.; Enewold, L.R.; Pfeiffer, R.M. The risk of amyotrophic lateral sclerosis after cancer in U.S. elderly adults: A population-based prospective study. Int. J. Cancer 2014, 135, 1745–1750, doi:10.1002/ijc.28795.
- Pan, T.; Li, X.; Jankovic, J. The association between Parkinson’s disease and melanoma. Int. J. Cancer 2011, 128, 2251–2260, doi:10.1002/ijc.25912.
- Hu, H.-H.; Kannengiesser, C.; Lesage, S.; André, J.; Mourah, S.; Michel, L.; Descamps, V.; Basset-Seguin, N.; Bagot, M.; Bensussan, A.; et al. PARKIN Inactivation Links Parkinson’s Disease to Melanoma. JNCI: J. Natl. Cancer Inst. 2015, 108, doi:10.1093/jnci/djv340.
- Kareus, S.A.; Figueroa, K.P.; Cannon-Albright, L.A.; Pulst, S.M. Shared predispositions of parkinsonism and cancer: A population-based pedigree-linked study. Arch. Neurol. 2012, 69, 1572–1577, doi:10.1001/archneurol.2012.2261.
- Liu, R.; Gao, X.; Lu, Y.; Chen, H. Meta-analysis of the relationship between Parkinson disease and melanoma. Neurology 2011, 76, 2002–2009, doi:10.1212/WNL.0b013e31821e554e.
- Walter, U.; Heilmann, E.; Voss, J.; Riedel, K.; Zhivov, A.; Schäd, S.G.; Gross, G.E.; Benecke, R.; Trcka, J. Frequency and profile of Parkinson’s disease prodromi in patients with malignant melanoma. J. Neurol. Neurosurg. Psychiatry 2016, 87, 302–310, doi:10.1136/jnnp-2014-310239.
- Frain, L.; Swanson, D.; Cho, K.; Gagnon, D.; Lu, K.P.; Betensky, R.A.; Driver, J. Association of cancer and Alzheimer’s disease risk in a national cohort of veterans. Alzheimer’s Dement. 2017, 13, 1364–1370, doi:10.1016/j.jalz.2017.04.012.
- Kesler, S.R.; Watson, C.L.; Blayney, D.W. Brain network alterations and vulnerability to simulated neurodegeneration in breast cancer. Neurobiol. Aging 2015, 36, 2429–2442, doi:10.1016/j.neurobiolaging.2015.04.015.
- Ganguli, M. Cancer and Dementia: It’s Complicated. Alzheimer Dis. Assoc. Disord. 2015, 29, 177–182, doi:10.1097/WAD.0000000000000086.
- Houck, A.L.; Seddighi, S.; Driver, J.A. At the Crossroads Between Neurodegeneration and Cancer: A Review of Overlapping Biology and Its Implications. Curr. Aging Sci. 2018, 11, 77–89, doi:10.2174/1874609811666180223154436.
- Benilova, I.; De Strooper, B. Prion protein in Alzheimer’s pathogenesis: A hot and controversial issue. EMBO Mol. Med. 2010, 2, 289–290, doi:10.1002/emmm.201000088.
- Driver, J.A. Inverse association between cancer and neurodegenerative disease: Review of the epidemiologic and biological evidence. Biogerontology 2014, 15, 547–557, doi:10.1007/s10522-014-9523-2.
- Du, L.; Pertsemlidis, A. Cancer and neurodegenerative disorders: Pathogenic convergence through microRNA regulation. J. Mol. Cell Biol. 2011, 3, 176–180, doi:10.1093/jmcb/mjq058.
- Klus, P.; Cirillo, D.; Botta Orfila, T.; Gaetano Tartaglia, G. Neurodegeneration and Cancer: Where the Disorder Prevails. Sci. Rep. 2015, 5, 15390–15390, doi:10.1038/srep15390.
- Plun-Favreau, H.; Lewis, P.A.; Hardy, J.; Martins, L.M.; Wood, N.W. Cancer and neurodegeneration: Between the devil and the deep blue sea. PLoS Genet. 2010, 6, e1001257, doi:10.1371/journal.pgen.1001257.
- Morris, L.G.T.; Veeriah, S.; Chan, T.A. Genetic determinants at the interface of cancer and neurodegenerative disease. Oncogene 2010, 29, 3453–3464, doi:10.1038/onc.2010.127.
- Feng, Y.-C.A.; Cho, K.; Lindstrom, S.; Kraft, P.; Cormack, J.; Blalock, K.; Campbell, P.T.; Casey, G.; Conti, D.V.; Edlund, C.K.; et al. Investigating the genetic relationship between Alzheimer’s disease and cancer using GWAS summary statistics. Hum. Genet. 2017, 136, 1341–1351, doi:10.1007/s00439-017-1831-6.
- Ibáñez, K.; Boullosa, C.; Tabarés-Seisdedos, R.; Baudot, A.; Valencia, A. Molecular evidence for the inverse comorbidity between central nervous system disorders and cancers detected by transcriptomic meta-analyses. PLoS Genet. 2014, 10, e1004173, doi:10.1371/journal.pgen.1004173.
- Wu, X.; Chen, S.; Lu, C. Amyloid precursor protein promotes the migration and invasion of breast cancer cells by regulating the MAPK signaling pathway. Int. J. Mol. Med. 2020, 45, 162–174, doi:10.3892/ijmm.2019.4404.
- Tsang, J.Y.S.; Lee, M.A.; Ni, Y.-B.; Chan, S.-K.; Cheung, S.-Y.; Chan, W.-W.; Lau, K.-F.; Tse, G.M.K. Amyloid Precursor Protein Is Associated with Aggressive Behavior in Nonluminal Breast Cancers. Oncologist 2018, 23, 1273–1281, doi:10.1634/theoncologist.2018-0012.
- Ge, Y.; Xu, K. Alpha-synuclein contributes to malignant progression of human meningioma via the Akt/mTOR pathway. Cancer Cell Int. 2016, 16, 86, doi:10.1186/s12935-016-0361-y.
- Rossi, G.; Redaelli, V.; Contiero, P.; Fabiano, S.; Tagliabue, G.; Perego, P.; Benussi, L.; Bruni, A.C.; Filippini, G.; Farinotti, M.; et al. Tau Mutations Serve as a Novel Risk Factor for Cancer. Cancer Res. 2018, 78, 3731–3739, doi:10.1158/0008-5472.can-17-3175.
- Rossi, G.; Conconi, D.; Panzeri, E.; Redaelli, S.; Piccoli, E.; Paoletta, L.; Dalprà, L.; Tagliavini, F. Mutations in MAPT gene cause chromosome instability and introduce copy number variations widely in the genome. J. Alzheimers Dis. 2013, 33, 969–982, doi:10.3233/jad-2012-121633.
- Buée, L.; Bussière, T.; Buée-Scherrer, V.; Delacourte, A.; Hof, P.R. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res. Rev. 2000, 33, 95–130, doi:10.1016/s0165-0173(00)00019-9.
- Goedert, M.; Spillantini, M.G.; Crowther, R.A. Cloning of a big tau microtubule-associated protein characteristic of the peripheral nervous system. Proc. Natl. Acad. Sci. USA 1992, 89, 1983–1987, doi:10.1073/pnas.89.5.1983.
- Andreadis, A. Tau gene alternative splicing: Expression patterns, regulation and modulation of function in normal brain and neurodegenerative diseases. Biochim. Biophys. Acta 2005, 1739, 91–103, doi:10.1016/j.bbadis.2004.08.010.
- Mandelkow, E.M.; Mandelkow, E. Biochemistry and cell biology of tau protein in neurofibrillary degeneration. Cold Spring Harbor Perspect. Med. 2012, 2, a006247, doi:10.1101/cshperspect.a006247.
- Tolnay; Probst. REVIEW: Tau protein pathology in Alzheimer’s disease and related disorders. Neuropathol. Appl. Neurobiol. 1999, 25, 171–187, doi:10.1046/j.1365-2990.1999.00182.x.
- Yoshida, M. Cellular tau pathology and immunohistochemical study of tau isoforms in sporadic tauopathies. Neuropathology 2006, 26, 457–470, doi:10.1111/j.1440-1789.2006.00743.x.
- Askanas, V.; Engel, W.K. Inclusion-body myositis: Muscle-fiber molecular pathology and possible pathogenic significance of its similarity to Alzheimer’s and Parkinson’s disease brains. Acta Neuropathol. 2008, 116, 583–595, doi:10.1007/s00401-008-0449-0.
- François, M.; Leifert, W.; Martins, R.; Thomas, P.; Fenech, M. Biomarkers of Alzheimer’s disease risk in peripheral tissues; focus on buccal cells. Curr. Alzheimer Res. 2014, 11, 519–531, doi:10.2174/1567205011666140618103827.
- Hattori, H.; Matsumoto, M.; Iwai, K.; Tsuchiya, H.; Miyauchi, E.; Takasaki, M.; Kamino, K.; Munehira, J.; Kimura, Y.; Kawanishi, K.; et al. The tau protein of oral epithelium increases in Alzheimer’s disease. J. Gerontol. Ser. A Biol. Sci. Med Sci. 2002, 57, M64–M70, doi:10.1093/gerona/57.1.m64.
- Ingelson, M.; Vanmechelen, E.; Lannfelt, L. Microtubule-associated protein tau in human fibroblasts with the Swedish Alzheimer mutation. Neurosci. Lett. 1996, 220, 9–12, doi:10.1016/s0304-3940(96)13218-3.
- Miklossy, J.; Qing, H.; Radenovic, A.; Kis, A.; Vileno, B.; Làszló, F.; Miller, L.; Martins, R.N.; Waeber, G.; Mooser, V.; et al. Beta amyloid and hyperphosphorylated tau deposits in the pancreas in type 2 diabetes. Neurobiol. Aging 2010, 31, 1503–1515, doi:10.1016/j.neurobiolaging.2008.08.019.
- Rouzier, R.; Rajan, R.; Wagner, P.; Hess, K.R.; Gold, D.L.; Stec, J.; Ayers, M.; Ross, J.S.; Zhang, P.; Buchholz, T.A.; et al. Microtubule-associated protein tau: A marker of paclitaxel sensitivity in breast cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 8315–8320, doi:10.1073/pnas.0408974102.
- Shi, M.; Sui, Y.-T.; Peskind, E.R.; Li, G.; Hwang, H.; Devic, I.; Ginghina, C.; Edgar, J.S.; Pan, C.; Goodlett, D.R.; et al. Salivary tau species are potential biomarkers of Alzheimer’s disease. J. Alzheimers Dis. 2011, 27, 299–305, doi:10.3233/JAD-2011-110731.
- Souter, S.; Lee, G. Microtubule-associated protein tau in human prostate cancer cells: Isoforms, phosphorylation, and interactions. J. Cell. Biochem. 2009, 108, 555–564, doi:10.1002/jcb.22287.
- Dugger, B.N.; Whiteside, C.M.; Maarouf, C.L.; Walker, D.G.; Beach, T.G.; Sue, L.I.; Garcia, A.; Dunckley, T.; Meechoovet, B.; Reiman, E.M.; et al. The Presence of Select Tau Species in Human Peripheral Tissues and Their Relation to Alzheimer’s Disease. J. Alzheimers Dis. 2016, 51, 345–356, doi:10.3233/jad-150859.
- Gu, Y.; Oyama, F.; Ihara, Y. τ Is Widely Expressed in Rat Tissues. J. Neurochem. 1996, 67, 1235–1244, doi:10.1046/j.1471-4159.1996.67031235.x.
- Boyne, L.J.; Tessler, A.; Murray, M.; Fischer, I. Distribution of Big tau in the central nervous system of the adult and developing rat. J. Comp. Neurol. 1995, 358, 279–293, doi:10.1002/cne.903580209.
- Couchie, D.; Mavilia, C.; Georgieff, I.S.; Liem, R.K.; Shelanski, M.L.; Nunez, J. Primary structure of high molecular weight tau present in the peripheral nervous system. Proc. Natl. Acad. Sci. USA 1992, 89, 4378–4381, doi:10.1073/pnas.89.10.4378.
- Lau, D.H.W.; Hogseth, M.; Phillips, E.C.; O’Neill, M.J.; Pooler, A.M.; Noble, W.; Hanger, D.P. Critical residues involved in tau binding to fyn: Implications for tau phosphorylation in Alzheimer’s disease. Acta Neuropathol. Commun. 2016, 4, 49, doi:10.1186/s40478-016-0317-4.
- Loomis, P.A.; Howard, T.H.; Castleberry, R.P.; Binder, L.I. Identification of nuclear tau isoforms in human neuroblastoma cells. Proc. Natl. Acad. Sci. USA 1990, 87, 8422–8426, doi:10.1073/pnas.87.21.8422.
- Wang, Y.; Loomis, P.A.; Zinkowski, R.P.; Binder, L.I. A novel tau transcript in cultured human neuroblastoma cells expressing nuclear tau. J. Cell Biol. 1993, 121, 257–267, doi:10.1083/jcb.121.2.257.
- Sultan, A.; Nesslany, F.; Violet, M.; Bégard, S.; Loyens, A.; Talahari, S.; Mansuroglu, Z.; Marzin, D.; Sergeant, N.; Humez, S.; et al. Nuclear Tau, a Key Player in Neuronal DNA Protection. J. Biol. Chem. 2011, 286, 4566–4575, doi:10.1074/jbc.M110.199976.
- Violet, M.; Delattre, L.; Tardivel, M.; Sultan, A.; Chauderlier, A.; Caillierez, R.; Talahari, S.; Nesslany, F.; Lefebvre, B.; Bonnefoy, E.; et al. A major role for Tau in neuronal DNA and RNA protection in vivo under physiological and hyperthermic conditions. Front. Cell. Neurosci. 2014, 8, 84, doi:10.3389/fncel.2014.00084.
- Frost, B.; Hemberg, M.; Lewis, J.; Feany, M.B. Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci. 2014, 17, 357–366, doi:10.1038/nn.3639.
- Eftekharzadeh, B.; Daigle, J.G.; Kapinos, L.E.; Coyne, A.; Schiantarelli, J.; Carlomagno, Y.; Cook, C.; Miller, S.J.; Dujardin, S.; Amaral, A.S.; et al. Tau Protein Disrupts Nucleocytoplasmic Transport in Alzheimer’s Disease. Neuron 2019, 101, 349, doi:10.1016/j.neuron.2018.12.031.
- Guo, C.; Jeong, H.-H.; Hsieh, Y.-C.; Klein, H.-U.; Bennett, D.A.; De Jager, P.L.; Liu, Z.; Shulman, J.M. Tau Activates Transposable Elements in Alzheimer’s Disease. Cell Rep. 2018, 23, 2874–2880, doi:10.1016/j.celrep.2018.05.004.
- Maina, M.B.; Bailey, L.J.; Wagih, S.; Biasetti, L.; Pollack, S.J.; Quinn, J.P.; Thorpe, J.R.; Doherty, A.J.; Serpell, L.C. The involvement of tau in nucleolar transcription and the stress response. Acta Neuropathol. Commun. 2018, 6, 70, doi:10.1186/s40478-018-0565-6.
- Sjöberg, M.K.; Shestakova, E.; Mansuroglu, Z.; Maccioni, R.B.; Bonnefoy, E. Tau protein binds to pericentromeric DNA: A putative role for nuclear tau in nucleolar organization. J. Cell Sci. 2006, 119, 2025–2034, doi:10.1242/jcs.02907.
- Sola, M.; Magrin, C.; Pedrioli, G.; Pinton, S.; Salvadè, A.; Papin, S.; Paganetti, P. Tau affects P53 function and cell fate during the DNA damage response. Commun. Biol. 2020, 3, 245, doi:10.1038/s42003-020-0975-4.
- Ferreira, C.G.; Tolis, C.; Giaccone, G. p53 and chemosensitivity. Ann. Oncol. 1999, 10, 1011–1022, doi:10.1023/A:1008361818480.
- Gradishar, W.J. Taxanes for the treatment of metastatic breast cancer. Breast cancer 2012, 6, 159–171, doi:10.4137/bcbcr.s8205.
- Greenberger, L.M.; Sampath, D. Resistance To Taxanes. In Cancer Drug Resistance, Teicher, B.A., Ed. Humana Press: Totowa, NJ, USA, 2006; pp. 329–358, doi:10.1007/978-1-59745-035-5_18.
- Kar, S.; Fan, J.; Smith, M.J.; Goedert, M.; Amos, L.A. Repeat motifs of tau bind to the insides of microtubules in the absence of taxol. EMBO J. 2003, 22, 70–77, doi:10.1093/emboj/cdg001.
- Ikeda, H.; Taira, N.; Hara, F.; Fujita, T.; Yamamoto, H.; Soh, J.; Toyooka, S.; Nogami, T.; Shien, T.; Doihara, H.; et al. The estrogen receptor influences microtubule-associated protein tau (MAPT) expression and the selective estrogen receptor inhibitor fulvestrant downregulates MAPT and increases the sensitivity to taxane in breast cancer cells. Breast Cancer Res. 2010, 12, R43, doi:10.1186/bcr2598.
- Smoter, M.; Bodnar, L.; Duchnowska, R.; Stec, R.; Grala, B.; Szczylik, C. The role of Tau protein in resistance to paclitaxel. Cancer Chemother. Pharmacol. 2011, 68, 553–557, doi:10.1007/s00280-011-1696-7.
- Wagner, P.; Wang, B.; Clark, E.; Lee, H.; Rouzier, R.; Pusztai, L. Microtubule Associated Protein (MAP)-Tau: A Novel Mediator of Paclitaxel Sensitivity In Vitro and In Vivo. Cell Cycle 2005, 4, 1149–1152, doi:10.4161/cc.4.9.2038.
- Smith, M.J.; Crowther, R.A.; Goedert, M. The natural osmolyte trimethylamine N-oxide (TMAO) restores the ability of mutant tau to promote microtubule assembly. FEBS Lett. 2000, 484, 265–270, doi:10.1016/s0014-5793(00)02169-4.
- Valet, F.; de Cremoux, P.; Spyratos, F.; Servant, N.; Dujaric, M.E.; Gentien, D.; Lehmann-Che, J.; Scott, V.; Sigal-Zafrani, B.; Mathieu, M.C.; et al. Challenging single- and multi-probesets gene expression signatures of pathological complete response to neoadjuvant chemotherapy in breast cancer: Experience of the REMAGUS 02 phase II trial. Breast 2013, 22, 1052–1059, doi:10.1016/j.breast.2013.08.015.
- Smoter, M.; Bodnar, L.; Grala, B.; Stec, R.; Zieniuk, K.; Kozlowski, W.; Szczylik, C. Tau protein as a potential predictive marker in epithelial ovarian cancer patients treated with paclitaxel/platinum first-line chemotherapy. J. Exp. Clin. Cancer Res. 2013, 32, 25, doi:10.1186/1756-9966-32-25.
- Yamauchi, A.; Kobayashi, A.; Oikiri, H.; Yokoyama, Y. Functional role of the Tau protein in epithelial ovarian cancer cells. Reprod. Med. Biol. 2017, 16, 143–151, doi:10.1002/rmb2.12019.
- Mimori, K.; Sadanaga, N.; Yoshikawa, Y.; Ishikawa, K.; Hashimoto, M.; Tanaka, F.; Sasaki, A.; Inoue, H.; Sugimachi, K.; Mori, M. Reduced tau expression in gastric cancer can identify candidates for successful Paclitaxel treatment. Br. J. Cancer 2006, 94, 1894–1897, doi:10.1038/sj.bjc.6603182.
- Sangrajrang, S.; Denoulet, P.; Millot, G.; Tatoud, R.; Podgorniak, M.P.; Tew, K.D.; Calvo, F.; Fellous, A. Estramustine resistance correlates with tau over-expression in human prostatic carcinoma cells. Int. J. Cancer 1998, 77, 626–631, doi:10.1002/(sici)1097-0215(19980812)77:4<626::aid-ijc24>3.0.co;2-2.
- Yoo, J.; Shim, B.Y.; Yoo, C.Y.; Kang, S.J.; Lee, K.Y. Predictive Significance of KRAS and Tau for Chemoresponse in Advanced Non-Small-Cell Lung Cancer. J. Pathol. Transl. Med. 2009, 43, 435–440, doi:10.4132/KoreanJPathol.2009.43.5.435.
- Guise, S.; Braguer, D.; Remacle-Bonnet, M.; Pommier, G.; Briand, C. Tau protein is involved in the apoptotic process induced by anti-microtubule agents on neuroblastoma cells. Apoptosis 1999, 4, 47–58, doi:10.1023/A:1009682116158.
- Baquero, M.T.; Lostritto, K.; Gustavson, M.D.; Bassi, K.A.; Appia, F.; Camp, R.L.; Molinaro, A.M.; Harris, L.N.; Rimm, D.L. Evaluation of prognostic and predictive value of microtubule associated protein tau in two independent cohorts. Breast Cancer Res. 2011, 13, R85, doi:10.1186/bcr2937.
- Pentheroudakis, G.; Kalogeras, K.T.; Wirtz, R.M.; Grimani, I.; Zografos, G.; Gogas, H.; Stropp, U.; Pectasides, D.; Skarlos, D.; Hennig, G.; et al. Gene expression of estrogen receptor, progesterone receptor and microtubule-associated protein Tau in high-risk early breast cancer: A quest for molecular predictors of treatment benefit in the context of a Hellenic Cooperative Oncology Group trial. Breast Cancer Res. Treat. 2009, 116, 131–143, doi:10.1007/s10549-008-0144-9.
- Pusztai, L.; Jeong, J.-H.; Gong, Y.; Ross, J.S.; Kim, C.; Paik, S.; Rouzier, R.; Andre, F.; Hortobagyi, G.N.; Wolmark, N.; et al. Evaluation of microtubule-associated protein-Tau expression as a prognostic and predictive marker in the NSABP-B 28 randomized clinical trial. J. Clin. Oncol. 2009, 27, 4287–4292, doi:10.1200/JCO.2008.21.6887.
- Yang, J.; Yu, Y.; Liu, W.; Li, Z.; Wei, Z.; Jiang, R. Microtubule-associated protein tau is associated with the resistance to docetaxel in prostate cancer cell lines. Res. Rep. Urol. 2017, 9, 71–77, doi:10.2147/RRU.S118966.
- Wu, H.; Huang, M.; Lu, M.; Zhu, W.; Shu, Y.; Cao, P.; Liu, P. Regulation of microtubule-associated protein tau (MAPT) by miR-34c-5p determines the chemosensitivity of gastric cancer to paclitaxel. Cancer Chemother. Pharmacol. 2013, 71, 1159–1171, doi:10.1007/s00280-013-2108-y.
- Ye, J.; Zhang, Z.; Sun, L.; Fang, Y.; Xu, X.; Zhou, G. miR-186 regulates chemo-sensitivity to paclitaxel via targeting MAPT in non-small cell lung cancer (NSCLC). Mol. Biosyst. 2016, 12, 3417–3424, doi:10.1039/c6mb00576d.
- Hristodorov, D.; Mladenov, R.; Pardo, A.; Pham, A.T.; Huhn, M.; Fischer, R.; Thepen, T.; Barth, S. Microtubule-associated protein tau facilitates the targeted killing of proliferating cancer cells in vitro and in a xenograft mouse tumour model in vivo. Br. J. Cancer 2013, 109, 1570–1578, doi:10.1038/bjc.2013.457.
- Akinrinmade, O.A.; Jordaan, S.; Hristodorov, D.; Mladenov, R.; Mungra, N.; Chetty, S.; Barth, S. Human MAP Tau Based Targeted Cytolytic Fusion Proteins. Biomedicines 2017, 5, 36, doi:10.3390/biomedicines5030036.
- Bonneau, C.; Gurard-Levin, Z.A.; Andre, F.; Pusztai, L.; Rouzier, R. Predictive and Prognostic Value of the TauProtein in Breast Cancer. Anticancer Res. 2015, 35, 5179–5184.
- Shao, Y.Y.; Kuo, K.T.; Hu, F.C.; Lu, Y.S.; Huang, C.S.; Liau, J.Y.; Lee, W.C.; Hsu, C.; Kuo, W.H.; Chang, K.J.; et al. Predictive and prognostic values of tau and ERCC1 in advanced breast cancer patients treated with paclitaxel and cisplatin. Jpn. J. Clin. Oncol. 2010, 40, 286–293, doi:10.1093/jjco/hyp184.
- Andre, F.; Hatzis, C.; Anderson, K.; Sotiriou, C.; Mazouni, C.; Mejia, J.; Wang, B.; Hortobagyi, G.N.; Symmans, W.F.; Pusztai, L. Microtubule-associated protein-tau is a bifunctional predictor of endocrine sensitivity and chemotherapy resistance in estrogen receptor-positive breast cancer. Clin. Cancer Res. 2007, 13, 2061–2067, doi:10.1158/1078-0432.ccr-06-2078.
- Ferreira, A.; Caceres, A. Estrogen-enhanced neurite growth: Evidence for a selective induction of Tau and stable microtubules. J. Neurosci. 1991, 11, 392–400, doi:10.1523/jneurosci.11-02-00392.1991.
- Frasor, J.; Stossi, F.; Danes, J.M.; Komm, B.; Lyttle, C.R.; Katzenellenbogen, B.S. Selective estrogen receptor modulators: Discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Res. 2004, 64, 1522–1533, doi:10.1158/0008-5472.can-03-3326.
- Lew, G.M. Changes in microtubular tau protein after estrogen in a cultured human neuroblastoma cell line. Gen. Pharmacol. 1993, 24, 1383–1386, doi:10.1016/0306-3623(93)90423-u.
- Ma, Z.Q.; Spreafico, E.; Pollio, G.; Santagati, S.; Conti, E.; Cattaneo, E.; Maggi, A. Activated estrogen receptor mediates growth arrest and differentiation of a neuroblastoma cell line. Proc. Natl. Acad. Sci. USA 1993, 90, 3740–3744, doi:10.1073/pnas.90.8.3740.
- Matsuno, A.; Takekoshi, S.; Sanno, N.; Utsunomiya, H.; Ohsugi, Y.; Saito, N.; Kanemitsu, H.; Tamura, A.; Nagashima, T.; Osamura, R.Y.; et al. Modulation of protein kinases and microtubule-associated proteins and changes in ultrastructure in female rat pituitary cells: Effects of estrogen and bromocriptine. J. Histochem. Cytochem. 1997, 45, 805–813, doi:10.1177/002215549704500605.
- West, M.; Blanchette, C.; Dressman, H.; Huang, E.; Ishida, S.; Spang, R.; Zuzan, H.; Olson, J.A.; Marks, J.R.; Nevins, J.R. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc. Natl. Acad. Sci. USA 2001, 98, 11462–11467, doi:10.1073/pnas.201162998.
- Osborne, C.K.; Wakeling, A.; Nicholson, R.I. Fulvestrant: An oestrogen receptor antagonist with a novel mechanism of action. Br. J. Cancer 2004, 90 (Suppl. 1), S2–S6, doi:10.1038/sj.bjc.6601629.
- Sui, M.; Huang, Y.; Park, B.H.; Davidson, N.E.; Fan, W. Estrogen receptor alpha mediates breast cancer cell resistance to paclitaxel through inhibition of apoptotic cell death. Cancer Res. 2007, 67, 5337–5344, doi:10.1158/0008-5472.can-06-4582.
- Gargini, R.; Segura-Collar, B.; Sánchez-Gómez, P. Novel Functions of the Neurodegenerative-Related Gene Tau in Cancer. Front. Aging Neurosci. 2019, 11, doi:10.3389/fnagi.2019.00231.
- Matrone, M.A.; Whipple, R.A.; Thompson, K.; Cho, E.H.; Vitolo, M.I.; Balzer, E.M.; Yoon, J.R.; Ioffe, O.B.; Tuttle, K.C.; Tan, M.; et al. Metastatic breast tumors express increased tau, which promotes microtentacle formation and the reattachment of detached breast tumor cells. Oncogene 2010, 29, 3217–3227, doi:10.1038/onc.2010.68.
- Cirak, Y.; Sarsik, B.; Cakar, B.; Sen, S.; Simsir, A.; Uslu, R. Predictive and prognostic values of Tau and BubR1 protein in prostate cancer and their relationship to the Gleason score. Med Oncol. 2013, 30, 526, doi:10.1007/s12032-013-0526-7.
- Schroeder, C.; Grell, J.; Hube-Magg, C.; Kluth, M.; Lang, D.; Simon, R.; Höflmayer, D.; Minner, S.; Burandt, E.; Clauditz, T.S.; et al. Aberrant expression of the microtubule-associated protein tau is an independent prognostic feature in prostate cancer. BMC Cancer 2019, 19, 193, doi:10.1186/s12885-019-5390-1.
- Tomlins, S.A.; Rhodes, D.R.; Perner, S.; Dhanasekaran, S.M.; Mehra, R.; Sun, X.W.; Varambally, S.; Cao, X.; Tchinda, J.; Kuefer, R.; et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005, 310, 644–648, doi:10.1126/science.1117679.
- Weischenfeldt, J.; Simon, R.; Feuerbach, L.; Schlangen, K.; Weichenhan, D.; Minner, S.; Wuttig, D.; Warnatz, H.J.; Stehr, H.; Rausch, T.; et al. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell 2013, 23, 159–170, doi:10.1016/j.ccr.2013.01.002.
- Birdsey, G.M.; Dryden, N.H.; Shah, A.V.; Hannah, R.; Hall, M.D.; Haskard, D.O.; Parsons, M.; Mason, J.C.; Zvelebil, M.; Gottgens, B.; et al. The transcription factor Erg regulates expression of histone deacetylase 6 and multiple pathways involved in endothelial cell migration and angiogenesis. Blood 2012, 119, 894–903, doi:10.1182/blood-2011-04-350025.
- Chow, A.; Amemiya, Y.; Sugar, L.; Nam, R.; Seth, A. Whole-transcriptome analysis reveals established and novel associations with TMPRSS2:ERG fusion in prostate cancer. Anticancer Res. 2012, 32, 3629–3641.
- Galletti, G.; Matov, A.; Beltran, H.; Fontugne, J.; Miguel Mosquera, J.; Cheung, C.; MacDonald, T.Y.; Sung, M.; O’Toole, S.; Kench, J.G.; et al. ERG induces taxane resistance in castration-resistant prostate cancer. Nat. Commun. 2014, 5, 5548, doi:10.1038/ncomms6548.
- Sekino, Y.; Han, X.; Babasaki, T.; Goto, K.; Inoue, S.; Hayashi, T.; Teishima, J.; Shiota, M.; Takeshima, Y.; Yasui, W.; et al. Microtubule-associated protein tau (MAPT) promotes bicalutamide resistance and is associated with survival in prostate cancer. Urol. Oncol. 2020, 38, 795.e791–795.e798, doi:10.1016/j.urolonc.2020.04.032.
- Smith, C.J.; Anderton, B.H.; Davis, D.R.; Gallo, J.M. Tau isoform expression and phosphorylation state during differentiation of cultured neuronal cells. FEBS Lett. 1995, 375, 243–248, doi:10.1016/0014-5793(95)01221-y.
- Tanaka, T.; Iqbal, K.; Trenkner, E.; Liu, D.J.; Grundke-Iqbal, I. Abnormally phosphorylated tau in SY5Y human neuroblastoma cells. FEBS Lett. 1995, 360, 5–9, doi:10.1016/0014-5793(95)00061-d.
- Reynolds, C.H.; Garwood, C.J.; Wray, S.; Price, C.; Kellie, S.; Perera, T.; Zvelebil, M.; Yang, A.; Sheppard, P.W.; Varndell, I.M.; et al. Phosphorylation regulates tau interactions with Src homology 3 domains of phosphatidylinositol 3-kinase, phospholipase Cgamma1, Grb2, and Src family kinases. J. Biol. Chem. 2008, 283, 18177–18186, doi:10.1074/jbc.M709715200.
- O’Neill, A.J.; Prencipe, M.; Dowling, C.; Fan, Y.; Mulrane, L.; Gallagher, W.M.; O’Connor, D.; O’Connor, R.; Devery, A.; Corcoran, C.; et al. Characterisation and manipulation of docetaxel resistant prostate cancer cell lines. Mol. Cancer 2011, 10, 126, doi:10.1186/1476-4598-10-126.
- Zaman, S.; Chobrutskiy, B.I.; Blanck, G. MAPT (Tau) expression is a biomarker for an increased rate of survival in pediatric neuroblastoma. Cell Cycle 2018, 17, 2474–2483, doi:10.1080/15384101.2018.1542898.
- Zaman, S.; Chobrutskiy, B.I.; Sikaria, D.; Blanck, G. MAPT (Tau) expression is a biomarker for an increased rate of survival for low‑grade glioma. Oncol. Rep. 2019, 41, 1359–1366, doi:10.3892/or.2018.6896.
- Gargini, R.; Segura-Collar, B.; Herránz, B.; García-Escudero, V.; Romero-Bravo, A.; Núñez, F.J.; García-Pérez, D.; Gutiérrez-Guamán, J.; Ayuso-Sacido, A.; Seoane, J.; et al. The IDH-TAU-EGFR triad defines the neovascular landscape of diffuse gliomas. Sci. Transl. Med. 2020, 12, doi:10.1126/scitranslmed.aax1501.
- Wang, C.; Liu, Y.; Guo, W.; Zhu, X.; Ahuja, N.; Fu, T. MAPT promoter CpG island hypermethylation is associated with poor prognosis in patients with stage II colorectal cancer. Cancer Manag. Res. 2019, 11, 7337–7343, doi:10.2147/cmar.s206731.
- Iwata, A.; Nagata, K.; Hatsuta, H.; Takuma, H.; Bundo, M.; Iwamoto, K.; Tamaoka, A.; Murayama, S.; Saido, T.; Tsuji, S. Altered CpG methylation in sporadic Alzheimer’s disease is associated with APP and MAPT dysregulation. Hum. Mol. Genet. 2014, 23, 648–656, doi:10.1093/hmg/ddt451.
- Coupland, K.G.; Mellick, G.D.; Silburn, P.A.; Mather, K.; Armstrong, N.J.; Sachdev, P.S.; Brodaty, H.; Huang, Y.; Halliday, G.M.; Hallupp, M.; et al. DNA methylation of the MAPT gene in Parkinson’s disease cohorts and modulation by vitamin E in vitro. Mov. Disord. 2014, 29, 1606–1614, doi:10.1002/mds.25784.
- Shui, I.M.; Wong, C.J.; Zhao, S.; Kolb, S.; Ebot, E.M.; Geybels, M.S.; Rubicz, R.; Wright, J.L.; Lin, D.W.; Klotzle, B.; et al. Prostate tumor DNA methylation is associated with cigarette smoking and adverse prostate cancer outcomes. Cancer 2016, 122, 2168–2177, doi:10.1002/cncr.30045.
- Kit, O.I.; Vodolazhsky, D.I.; Kutilin, D.S.; Enin, Y.S.; Gevorkyan, Y.A.; Zolotukhin, P.V.; Boumber, Y.; Kharin, L.V.; Panina, S.B. A Proteomics Analysis Reveals 9 Up-Regulated Proteins Associated with Altered Cell Signaling in Colon Cancer Patients. Protein J. 2017, 36, 513–522, doi:10.1007/s10930-017-9746-6.
- Huda, M.N.; Erdene-Ochir, E.; Pan, C.H. Assay for Phosphorylation and Microtubule Binding Along with Localization of Tau Protein in Colorectal Cancer Cells. J. Vis. Exp. 2017, doi:10.3791/55932.
- Bou Samra, E.; Buhagiar-Labarchède, G.; Machon, C.; Guitton, J.; Onclercq-Delic, R.; Green, M.R.; Alibert, O.; Gazin, C.; Veaute, X.; Amor-Guéret, M. A role for Tau protein in maintaining ribosomal DNA stability and cytidine deaminase-deficient cell survival. Nat. Commun. 2017, 8, 693, doi:10.1038/s41467-017-00633-1.