Cellular models have created opportunities to explore the characteristics of human diseases through well-established protocols, while avoiding the ethical restrictions associated with post-mortem studies and the costs associated with researching animal models. Organoids are 3D cellular structures that mimic the architecture and function of native tissues. They are generated in vitro from stem cells or differentiated cells, such as epithelial or neural cells, and are used to study organ development, disease modeling, and drug discovery. Organoids have become a powerful tool for understanding the cellular and molecular mechanisms underlying human physiology, providing new insights into the pathogenesis of cancer, metabolic diseases, and brain disorders.
- cell culture
- iPSCs
- disease modeling
- organoids
- 3D
- 2D
- cellular systems
1. Introduction
2. iPSCs
-
Ethical concerns: iPSCs can be derived from adult cells, avoiding the ethical concerns associated with embryonic stem cell research [30];
-
Patient-specific cells: iPSCs can be generated from a patient’s own cells, allowing for the creation of patient-specific cells for use in therapy. This can help to avoid immune rejection of the transplanted cells [31];
-
Disease modeling: iPSCs can be used to create models of specific diseases, which can aid in the understanding of the disease and the development of new treatments [18];
-
Tissue repair and regeneration: iPSCs have the potential to be used to repair or regenerate damaged or diseased tissue, such as in the treatment of heart disease, diabetes, and neurodegenerative disorders [15];
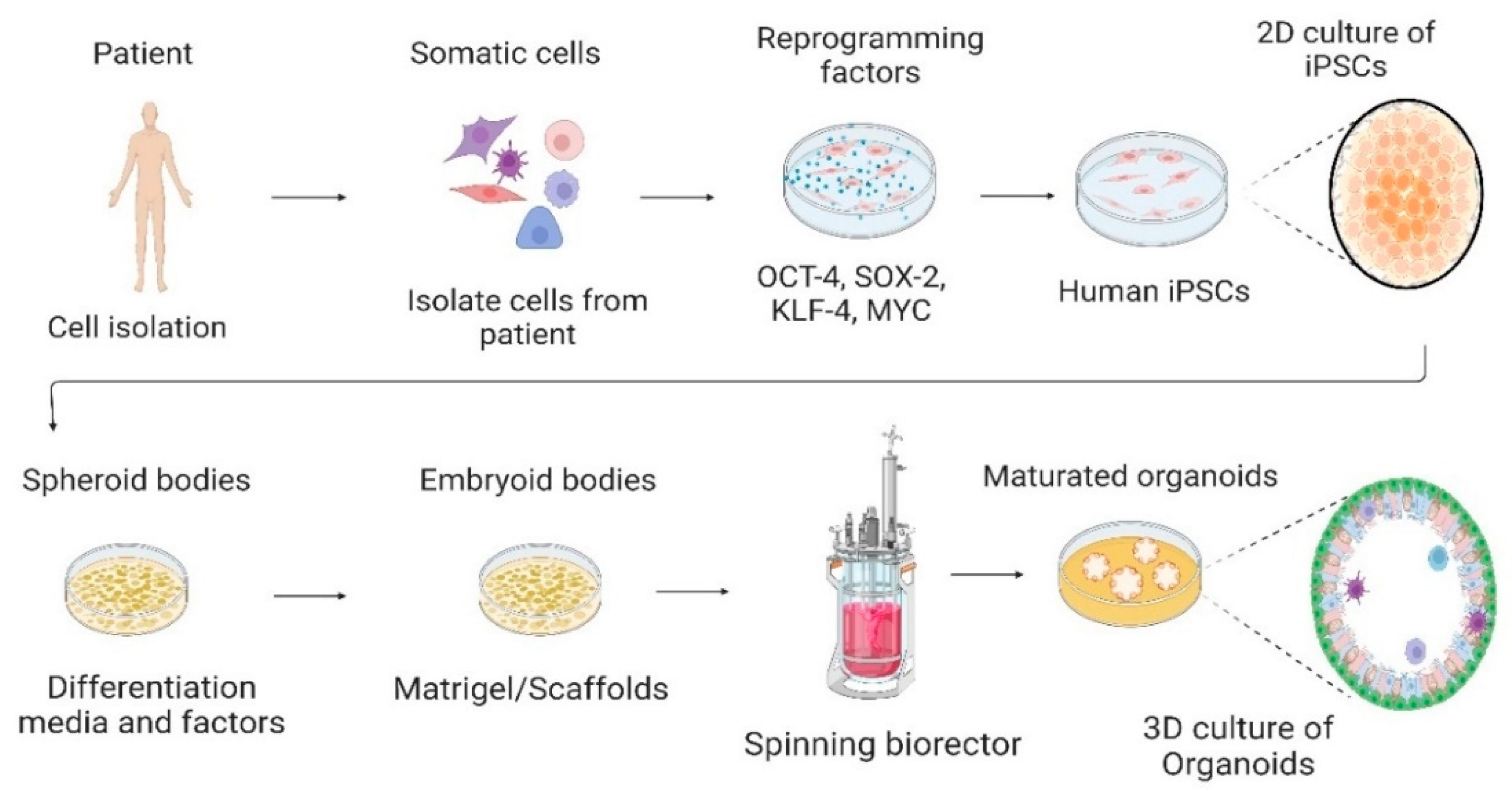
3. Cell Culture System—From 2D to 3D
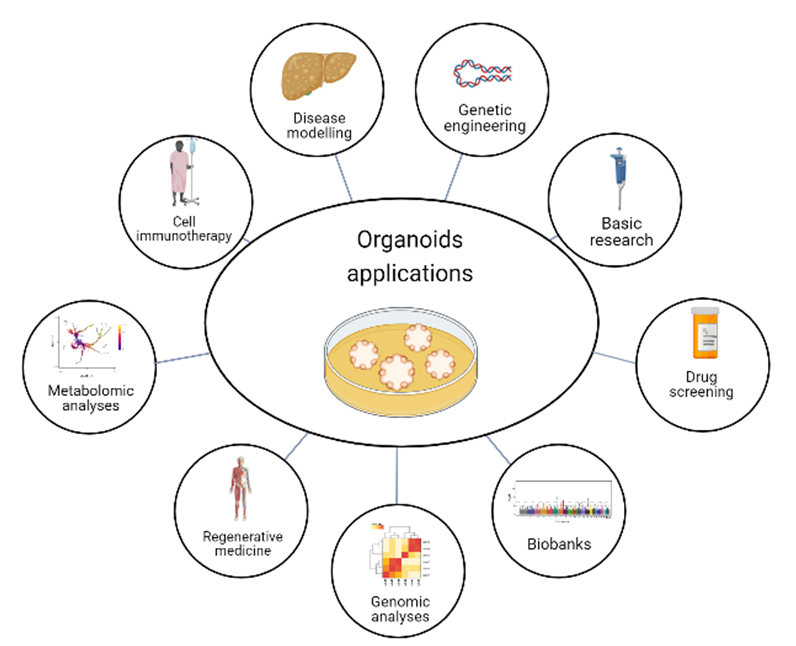
4. Organoids Technology
This entry is adapted from the peer-reviewed paper 10.3390/cells12060930
References
- Tidball, A.M. Disease in a Dish: Cellular Models to Understand Human Conditions. In Cellular and Animal Models in Human Genomics Research; Academic Press: Cambridge, MA, USA, 2019; pp. 19–47.
- Penney, J.; Ralvenius, W.T.; Tsai, L.H. Modeling Alzheimer’s disease with iPSC-derived brain cells. Mol. Psychiatry 2019, 25, 148–167.
- Avior, Y.; Sagi, I.; Benvenisty, N. Pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Mol. Cell Biol. 2016, 17, 170–182.
- Lancaster, M.A.; Huch, M. Disease modelling in human organoids. Dis. Model. Mech. 2019, 12, dmm039347.
- Schlaermann, P.; Toelle, B.; Berger, H.; Schmidt, S.C.; Glanemann, M.; Ordemann, J.; Bartfeld, S.; Mollenkopf, H.J.; Meyer, T.F. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut 2016, 65, 202–213.
- Heo, I.; Dutta, D.; Schaefer, D.A.; Iakobachvili, N.; Artegiani, B.; Sachs, N.; Boonekamp, K.E.; Bowden, G.; Hendrickx, A.P.A.; Willems, R.J.L.; et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 2018, 3, 814–823.
- Qian, X.; Nguyen, H.N.; Song, M.M.; Hadiono, C.; Ogden, S.C.; Hammack, C.; Yao, B.; Hamersky, G.R.; Jacob, F.; Zhong, C.; et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 2016, 165, 1238–1254.
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676.
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872.
- Nishikawa, S.-I.; Goldstein, R.A.; Nierras, C.R. The promise of human induced pluripotent stem cells for research and therapy. Nat. Rev. Mol. Cell Biol. 2008, 9, 725–729.
- Sürün, D.; Schneider, A.; Mircetic, J.; Neumann, K.; Lansing, F.; Paszkowski-Rogacz, M.; Hänchen, V.; Lee-Kirsch, M.A.; Buchholz, F. Efficient Generation and Correction of Mutations in Human iPS Cells Utilizing mRNAs of CRISPR Base Editors and Prime Editors. Genes 2020, 11, 511.
- Robinton, D.A.; Daley, G.Q. The promise of induced pluripotent stem cells in research and therapy. Nature 2012, 481, 295–305.
- Wen, Z.; Christian, K.M.; Song, H.; Ming, G.-L. Modeling psychiatric disorders with patient-derived iPSCs. Curr. Opin. Neurobiol. 2016, 36, 118–127.
- Brennand, K.; Simone, A.; Jou, J.; Gelboin-Burkhart, C.; Tran, N.; Sangar, S.; Li, Y.; Mu, Y.; Chen, G.; Yu, D.; et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 2011, 473, 221–225.
- Crook, J.M.; Tomaskovic-Crook, E. Bioprinting 3D Human Induced Pluripotent Stem Cell Constructs for Multilineage Tissue Engineering and Modeling. Methods Mol. Biol. 2020, 2140, 251–258.
- Csobonyeiova, M.; Polak, S.; Danisovic, L. Recent Overview of the Use of iPSCs Huntington’s Disease Modeling and Therapy. Int. J. Mol. Sci. 2020, 21, 2239.
- Karagiannis, P.; Takahashi, K.; Saito, M.; Yoshida, Y.; Okita, K.; Watanabe, A.; Inoue, H.; Yamashita, J.K.; Todani, M.; Nakagawa, M.; et al. Induced Pluripotent Stem Cells and Their Use in Human Models of Disease and Development. Physiol. Rev. 2019, 99, 79–114.
- Rowe, R.G.; Daley, G.Q. Induced pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Genet. 2019, 20, 377–388.
- Cavalli, E.; Battaglia, G.; Basile, M.S.; Bruno, V.; Petralia, M.C.; Lombardo, S.D.; Pennisi, M.; Kalfin, R.; Tancheva, L.; Fagone, P.; et al. Exploratory Analysis of iPSCS-Derived Neuronal Cells as Predictors of Diagnosis and Treatment of Alzheimer Disease. Brain Sci. 2020, 10, 166.
- Klotz, C.; Aebischer, T.; Seeber, F. Stem cell-derived cell cultures and organoids for protozoan parasite propagation and studying host–parasite interaction. Int. J. Med. Microbiol. 2012, 302, 203–209.
- Wang, Z.; Wang, S.-N.; Xu, T.-Y.; Miao, Z.-W.; Su, D.-F.; Miao, C.-Y. Organoid technology for brain and therapeutics research. CNS Neurosci. Ther. 2017, 23, 771–778.
- Shariati, L.; Esmaeili, Y.; Javanmard, S.H.; Bidram, E.; Amini, A. Organoid technology: Current standing and future perspectives. Stem Cells 2021, 39, 1625–1649.
- Dutta, D.; Heo, I.; Clevers, H. Disease Modeling in Stem Cell-Derived 3D Organoid Systems. Trends Mol. Med. 2017, 23, 393–410.
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147.
- Lee, S.H.; Lee, H.S.; Moon, H.C.; Kim, D.H.; Park, Y.S.; Hwang, B.; Lee, H.Y. The Effect of α-pinene from Pinus densiflora S. and a Polysaccharide from Angelica gigas Nakai on Differentiation and Proliferation of Human Embryonic Stem Cells. Cytotechnology 2005, 49, 87–94.
- Volarevic, V.; Markovic, B.S.; Gazdic, M.; Volarevic, A.; Jovicic, N.; Arsenijevic, N.; Armstrong, L.; Djonov, V.; Lako, M.; Stojkovic, M. Ethical and Safety Issues of Stem Cell-Based Therapy. Int. J. Med Sci. 2018, 15, 36–45.
- Zacharias, D.G.; Nelson, T.J.; Mueller, P.S.; Hook, C.C. The Science and Ethics of Induced Pluripotency: What Will Become of Embryonic Stem Cells? Mayo Clin. Proc. 2011, 86, 634–640.
- Deng, X.-Y.; Wang, H.; Wang, T.; Fang, X.-T.; Zou, L.; Li, Z.-Y.; Liu, C.-B. Non-Viral Methods For Generating Integration-Free, Induced Pluripotent Stem Cells. Curr. Stem Cell Res. Ther. 2015, 10, 153–158.
- Trevisan, M.; Desole, G.; Costanzi, G.; Lavezzo, E.; Palù, G.; Barzon, L. Reprogramming Methods Do Not Affect Gene Expression Profile of Human Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2017, 18, 206.
- Moradi, S.; Mahdizadeh, H.; Šarić, T.; Kim, J.; Harati, J.; Shahsavarani, H.; Greber, B.; Moore, J.B., IV. Research and therapy with induced pluripotent stem cells (iPSCs): Social, legal, and ethical considerations. Stem Cell Res. Ther. 2019, 10, 341.
- Paik, D.T.; Chandy, M.; Wu, J.C. Patient and Disease–Specific Induced Pluripotent Stem Cells for Discovery of Personalized Cardiovascular Drugs and Therapeutics. Pharmacol. Rev. 2019, 72, 320–342.
- Ovics, P.; Regev, D.; Baskin, P.; Davidor, M.; Shemer, Y.; Neeman, S.; Ben-Haim, Y.; Binah, O. Drug Development and the Use of Induced Pluripotent Stem Cell-Derived Cardiomyocytes for Disease Modeling and Drug Toxicity Screening. Int. J. Mol. Sci. 2020, 21, 7320.
- Pasteuning-Vuhman, S.; de Jongh, R.; Timmers, A.; Pasterkamp, R.J. Towards Advanced iPSC-based Drug Development for Neurodegenerative Disease. Trends Mol. Med. 2021, 27, 263–279.
- Lyra-Leite, D.M.; Fonoudi, H.; Gharib, M.; Burridge, P.W. An updated protocol for the cost-effective and weekend-free culture of human induced pluripotent stem cells. STAR Protoc. 2021, 2, 100213.
- D’Antonio, M.; Woodruff, G.; Nathanson, J.L.; D’Antonio-Chronowska, A.; Arias, A.; Matsui, H.; Williams, R.; Herrera, C.; Reyna, S.M.; Yeo, G.W.; et al. High-Throughput and Cost-Effective Characterization of Induced Pluripotent Stem Cells. Stem Cell Rep. 2017, 8, 1101–1111.
- Beers, J.; Linask, K.L.; Chen, J.A.; Siniscalchi, L.I.; Lin, Y.; Zheng, W.; Rao, M.; Chen, G. A cost-effective and efficient reprogramming platform for large-scale production of integration-free human induced pluripotent stem cells in chemically defined culture. Sci. Rep. 2015, 5, 11319.
- Csöbönyeiová, M.; Polák, Š.; Danišovič, L. Perspectives of induced pluripotent stem cells for cardiovascular system regeneration. Exp. Biol. Med. 2015, 240, 549–556.
- Rikhtegar, R.; Pezeshkian, M.; Dolati, S.; Safaie, N.; Rad, A.A.; Mahdipour, M.; Nouri, M.; Jodati, A.R.; Yousefi, M. Stem cells as therapy for heart disease: iPSCs, ESCs, CSCs, and skeletal myoblasts. Biomed. Pharmacother. 2019, 109, 304–313.
- Zhu, D.; Cheng, K. Cardiac Cell Therapy for Heart Repair: Should the Cells Be Left Out? Cells 2021, 10, 641.
- Walczak, M.P.; Drozd, A.M.; Stoczynska-Fidelus, E.; Rieske, P.; Grzela, D.P. Directed differentiation of human iPSC into insulin producing cells is improved by induced expression of PDX1 and NKX6.1 factors in IPC progenitors. J. Transl. Med. 2016, 14, 341.
- Silva, I.B.B.; Kimura, C.H.; Colantoni, V.P.; Sogayar, M.C. Stem cells differentiation into insulin-producing cells (IPCs): Recent advances and current challenges. Stem Cell Res. Ther. 2022, 13, 309.
- Poorna, M.; Sudhindran, S.; Thampi, M.; Mony, U. Differentiation of induced pluripotent stem cells to hepatocyte-like cells on cellulose nanofibril substrate. Colloids Surf. B Biointerfaces 2021, 198, 111466.
- Hu, C.; Li, L. In vitro culture of isolated primary hepatocytes and stem cell-derived hepatocyte-like cells for liver regeneration. Protein Cell 2015, 6, 562–574.
- Corbett, J.L.; Duncan, S.A. iPSC-Derived Hepatocytes as a Platform for Disease Modeling and Drug Discovery. Front. Med. 2019, 6, 265.
- Davidson, M.D.; Ware, B.R.; Khetani, S.R. Stem Cell-Derived Liver Cells for Drug Testing and Disease Modeling. Discov. Med. 2015, 19, 349–358.
- Nicholson, M.W.; Ting, C.Y.; Chan, D.Z.; Cheng, Y.C.; Lee, Y.C.; Hsu, C.C.; Huang, C.Y.; Hsieh, P.C. Utility of iPSC-Derived Cells for Disease Modeling, Drug Development, and Cell Therapy. Cells 2022, 11, 1853.
- Farkhondeh, A.; Li, R.; Gorshkov, K.; Chen, K.G.; Might, M.; Rodems, S.; Lo, D.C.; Zheng, W. Induced pluripotent stem cells for neural drug discovery. Drug Discov. Today 2019, 24, 992–999.
- Sharma, A.; McKeithan, W.L.; Serrano, R.; Kitani, T.; Burridge, P.W.; del Alamo, J.C.; Mercola, M.; Wu, J.C. Use of human induced pluripotent stem cell–derived cardiomyocytes to assess drug cardiotoxicity. Nat. Protoc. 2018, 13, 3018–3041.
- Ferreira, L.M.; Mostajo-Radji, M.A. How induced pluripotent stem cells are redefining personalized medicine. Gene 2013, 520, 1–6.
- Chandrasekaran, A.; Avci, H.X.; Ochalek, A.; Rösingh, L.N.; Molnár, K.; László, L.; Bellák, T.; Téglási, A.; Pesti, K.; Mike, A.; et al. Comparison of 2D and 3D neural induction methods for the generation of neural progenitor cells from human induced pluripotent stem cells. Stem Cell Res. 2017, 25, 139–151.
- Liu, C.; Oikonomopoulos, A.; Sayed, N.; Wu, J.C. Modeling human diseases with induced pluripotent stem cells: From 2D to 3D and beyond. Development 2018, 145, dev156166.
- Bordoni, M.; Rey, F.; Fantini, V.; Pansarasa, O.; Di Giulio, A.M.; Carelli, S.; Cereda, C. From Neuronal Differentiation of iPSCs to 3D Neuro-Organoids: Modelling and Therapy of Neurodegenerative Diseases. Int. J. Mol. Sci. 2018, 19, 3972.
- Costamagna, G.; Andreoli, L.; Corti, S.; Faravelli, I. iPSCs-Based Neural 3D Systems: A Multidimensional Approach for Disease Modeling and Drug Discovery. Cells 2019, 8, 1438.
- Caiazza, M.C.; Lang, C.; Wade-Martins, R. What we can learn from iPSC-derived cellular models of Parkinson’s disease. Prog. Brain Res. 2020, 252, 3–25.
- Torrent, R.; Rigotti, F.D.A.; Dell’Era, P.; Memo, M.; Raya, A.; Consiglio, A. Using iPS Cells toward the Understanding of Parkinson’s Disease. J. Clin. Med. 2015, 4, 548–566.
- Arber, C.; Toombs, J.; Lovejoy, C.; Ryan, N.S.; Paterson, R.W.; Willumsen, N.; Gkanatsiou, E.; Portelius, E.; Blennow, K.; Heslegrave, A.; et al. Familial Alzheimer’s disease patient-derived neurons reveal distinct mutation-specific effects on amyloid beta. Mol. Psychiatry 2019, 25, 2919–2931.
- Machairaki, V. Human Pluripotent Stem Cells as In Vitro Models of Neurodegenerative Diseases. Adv. Exp. Med. Biol. 2020, 1195, 93–94.
- Tcw, J. Human iPSC application in Alzheimer’s disease and Tau-related neurodegenerative diseases. Neurosci. Lett. 2019, 699, 31–40.
- Ellison, B. Stem Cell Research and Social Justice: Aligning Scientific Progress with Social Need. Curr. Stem Cell Rep. 2016, 2, 328–335.
- Ghosh, S.; Nehme, R.; Barrett, L.E. Greater genetic diversity is needed in human pluripotent stem cell models. Nat. Commun. 2022, 13, 7301.
- Omole, A.E.; Fakoya, A.O.J.; Nnawuba, K.C.; Haider, K.H. Common Ethical Considerations of Human-Induced Pluripotent. In Handbook of Stem Cell Therapy; Singapore: Singapore, 2022; pp. 1–17.
- Ayala, F.J. Cloning humans? Biological, ethical, and social considerations. Proc. Natl. Acad. Sci. USA 2015, 112, 8879–8886.
- Chlebanowska, P.; Sułkowski, M.; Skrzypek, K.; Tejchman, A.; Muszyńska, A.; Noroozi, R.; Majka, M. Origin of the Induced Pluripotent Stem Cells Affects Their Differentiation into Dopaminergic Neurons. Int. J. Mol. Sci. 2020, 21, 5705.
- Carvalho, A.B.; Coutinho, K.C.D.S.; Barbosa, R.A.Q.; de Campos, D.B.P.; Leitão, I.D.C.; Pinto, R.S.; Dos Santos, D.S.; Farjun, B.; Araújo, D.d.S.d.; Mesquita, F.C.P.; et al. Action potential variability in human pluripotent stem cell-derived cardiomyocytes obtained from healthy donors. Front. Physiol. 2022, 13, 2667.
- Féraud, O.; Valogne, Y.; Melkus, M.W.; Zhang, Y.; Oudrhiri, N.; Haddad, R.; Daury, A.; Rocher, C.; Larbi, A.; Duquesnoy, P.; et al. Donor Dependent Variations in Hematopoietic Differentiation among Embryonic and Induced Pluripotent Stem Cell Lines. PLoS ONE 2016, 11, e0149291.
- Poetsch, M.S.; Strano, A.; Guan, K. Human Induced Pluripotent Stem Cells: From Cell Origin, Genomic Stability, and Epigenetic Memory to Translational Medicine. Stem Cells 2022, 40, 546–555.
- Turinetto, V.; Orlando, L.; Giachino, C. Induced Pluripotent Stem Cells: Advances in the Quest for Genetic Stability during Reprogramming Process. Int. J. Mol. Sci. 2017, 18, 1952.
- Efrat, S. Epigenetic Memory: Lessons From iPS Cells Derived From Human β Cells. Front. Endocrinol. 2021, 11, 1063.
- Bar, S.; Benvenisty, N. Epigenetic aberrations in human pluripotent stem cells. EMBO J. 2019, 38, e101033.
- Sacco, A.M.; Belviso, I.; Romano, V.; Carfora, A.; Schonauer, F.; Nurzynska, D.; Montagnani, S.; Di Meglio, F.; Castaldo, C. Diversity of dermal fibroblasts as major determinant of variability in cell reprogramming. J. Cell. Mol. Med. 2019, 23, 4256–4268.
- Ortmann, D.; Vallier, L. Variability of human pluripotent stem cell lines. Curr. Opin. Genet. Dev. 2017, 46, 179–185.
- Chang, C.Y.; Ting, H.C.; Liu, C.A.; Su, H.L.; Chiou, T.W.; Harn, H.J.; Lin, S.Z.; Ho, T.J. Differentiation of Human Pluripotent Stem Cells Into Specific Neural Lineages. Cell Transplant. 2021, 30, 09636897211017829.
- Rouhani, F.J.; Zou, X.; Danecek, P.; Badja, C.; Amarante, T.D.; Koh, G.; Wu, Q.; Memari, Y.; Durbin, R.; Martincorena, I.; et al. Substantial somatic genomic variation and selection for BCOR mutations in human induced pluripotent stem cells. Nat. Genet. 2022, 54, 1406–1416.
- Lodrini, A.M.; Barile, L.; Rocchetti, M.; Altomare, C. Human Induced Pluripotent Stem Cells Derived from a Cardiac Somatic Source: Insights for an In-Vitro Cardiomyocyte Platform. Int. J. Mol. Sci. 2020, 21, 507.
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced pluripotent stem cell technology: A decade of progress. Nat. Rev. Drug Discov. 2017, 16, 115–130.
- Logan, S.; Arzua, T.; Canfield, S.G.; Seminary, E.R.; Sison, S.L.; Ebert, A.D.; Bai, X. Studying Human Neurological Disorders Using Induced Pluripotent Stem Cells: From 2D Monolayer to 3D Organoid and Blood Brain Barrier Models. Compr. Physiol. 2019, 9, 565–611.
- Hirschi, K.K.; Li, S.; Roy, K. Induced pluripotent stem cells for regenerative medicine. Annu. Rev. Biomed. Eng. 2014, 16, 277–294.
- Chang, N.C. Autophagy and Stem Cells: Self-Eating for Self-Renewal. Front. Cell Dev. Biol. 2020, 8, 138.
- Lee, C.-T.; Bendriem, R.M.; Wu, W.W.; Shen, R.-F. 3D brain Organoids derived from pluripotent stem cells: Promising experimental models for brain development and neurodegenerative disorders. J. Biomed. Sci. 2017, 24, 59.
- Mahla, R.S. Stem Cells Applications in Regenerative Medicine and Disease Therapeutics. Int. J. Cell Biol. 2016, 2016, 6940283.
- Suman, S.; Domingues, A.; Ratajczak, J.; Ratajczak, M.Z. Potential Clinical Applications of Stem Cells in Regenerative Medicine. Adv. Exp. Med. Biol. 2019, 1201, 1–22.
- Li, S.; Yang, K.; Chen, X.; Zhu, X.; Zhou, H.; Li, P.; Chen, Y.; Jiang, Y.; Li, T.; Qin, X.; et al. Simultaneous 2D and 3D cell culture array for multicellular geometry, drug discovery and tumor microenvironment reconstruction. Biofabrication 2021, 13, 045013.
- Muguruma, M.; Teraoka, S.; Miyahara, K.; Ueda, A.; Asaoka, M.; Okazaki, M.; Kawate, T.; Kuroda, M.; Miyagi, Y.; Ishikawa, T. Differences in drug sensitivity between two-dimensional and three-dimensional culture systems in triple-negative breast cancer cell lines. Biochem. Biophys. Res. Commun. 2020, 533, 268–274.
- Chaubey, A.; Ross, K.J.; Leadbetter, R.M.; Burg, K.J.L. Surface patterning: Tool to modulate stem cell differentiation in an adipose system. J. Biomed. Mater. Res. Part B Appl. Biomater. 2008, 84, 70–78.
- Mabry, K.M.; Payne, S.Z.; Anseth, K.S. Microarray analyses to quantify advantages of 2D and 3D hydrogel culture systems in maintaining the native valvular interstitial cell phenotype. Biomaterials 2016, 74, 31–41.
- Xu, X.; Wang, W.; Kratz, K.; Fang, L.; Li, Z.; Kurtz, A.; Ma, N.; Lendlein, A. Controlling major cellular processes of human mesenchymal stem cells using microwell structures. Adv. Healthc. Mater. 2014, 3, 1991–2003.
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33.
- Crosnier, C.; Staudt, N.; Wright, G.J. A rapid and scalable method for selecting recombinant mouse monoclonal antibodies. BMC Biol. 2010, 8, 76.
- Carrillo-Cocom, L.M.; Genel-Rey, T.; Araíz-Hernández, D.; López-Pacheco, F.; López-Meza, J.; Rocha-Pizaña, M.D.R.; Ramírez-Medrano, A.; Alvarez, M.M. Amino acid consumption in naïve and recombinant CHO cell cultures: Producers of a monoclonal antibody. Cytotechnology 2015, 67, 809–820.
- Jung-Klawitter, S.; Opladen, T. Induced pluripotent stem cells (iPSCs) as model to study inherited defects of neurotransmission in inborn errors of metabolism. J. Inherit. Metab. Dis. 2018, 41, 1103–1116.
- Mitra, A.; Mishra, L.; Li, S. Technologies for deriving primary tumor cells for use in personalized cancer therapy. Trends Biotechnol. 2013, 31, 347–354.
- Hsieh, C.-F.; Yan, Z.; Schumann, R.G.; Milz, S.; Pfeifer, C.G.; Schieker, M.; Docheva, D. In Vitro Comparison of 2D-Cell Culture and 3D-Cell Sheets of Scleraxis-Programmed Bone Marrow Derived Mesenchymal Stem Cells to Primary Tendon Stem/Progenitor Cells for Tendon Repair. Int. J. Mol. Sci. 2018, 19, 2272.
- Jin, L.; Qu, Y.; Gomez, L.J.; Chung, S.; Han, B.; Gao, B.; Yue, Y.; Gong, Y.; Liu, X.; Amersi, F.; et al. Characterization of primary human mammary epithelial cells isolated and propagated by conditional reprogrammed cell culture. Oncotarget 2018, 9, 11503–11514.
- Yin, Y.; Zhou, D. Organoid and enteroid modeling of Salmonella Infection. Front. Cell. Infect. Microbiol. 2018, 8, 102.
- Poon, A.; Zhang, Y.; Chandrasekaran, A.; Phanthong, P.; Schmid, B.; Nielsen, T.T.; Freude, K.K. Modeling neurodegenerative diseases with patient-derived induced pluripotent cells: Possibilities and challenges. New Biotechnol. 2017, 39, 190–198.
- Townsley, K.G.; Brennand, K.J.; Huckins, L.M. Massively parallel techniques for cataloguing the regulome of the human brain. Nat. Neurosci. 2020, 23, 1509–1521.
- Baxter, M.; Withey, S.; Harrison, S.; Segeritz, C.-P.; Zhang, F.; Atkinson-Dell, R.; Rowena, S.-Y.; Gerrard, D.T.; Sison-Young, R.; Jenkins, R.; et al. Phenotypic and functional analyses show stem cell-derived hepatocyte-like cells better mimic fetal rather than adult hepatocytes. J. Hepatol. 2015, 62, 581–589.
- Fontoura, J.C.; Viezzer, C.; dos Santos, F.G.; Ligabue, R.A.; Weinlich, R.; Puga, R.D.; Antonow, D.; Severino, P.; Bonorino, C. Comparison of 2D and 3D cell culture models for cell growth, gene expression and drug resistance. Mater. Sci. Eng. C 2020, 107, 110264.
- Li, D.-W.; He, F.-L.; He, J.; Deng, X.; Liu, Y.-L.; Liu, Y.-Y.; Ye, Y.-J.; Yin, D.-C. From 2D to 3D: The morphology, proliferation and differentiation of MC3T3-E1 on silk fibroin/chitosan matrices. Carbohydr. Polym. 2017, 178, 69–77.
- Langhans, S.A. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 2018, 9, 6.
- Paşca, A.M.; Sloan, S.A.; Clarke, L.E.; Tian, Y.; Makinson, C.D.; Huber, N.; Kim, C.H.; Park, J.-Y.; O’Rourke, N.A.; Nguyen, K.D.; et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 2015, 12, 671–678.
- Padmalayam, I.; Suto, M.J. 3D Cell Cultures: Mimicking In Vivo Tissues for Improved Predictability in Drug Discovery. Annu. Rep. Med. Chem. 2012, 47, 367–378.
- Zeng, Y.; Win-Shwe, T.-T.; Ito, T.; Sone, H. A three-dimensional neurosphere system using human stem cells for nanotoxicology studies. In Organoids and Mini-Organs; Academic Press: Cambridge, MA, USA, 2018; pp. 215–226.
- Yang, H.; Shao, N.; Holmström, A.; Zhao, X.; Chour, T.; Chen, H.; Itzhaki, I.; Wu, H.; Ameen, M.; Cunningham, N.J.; et al. Transcriptome analysis of non human primate-induced pluripotent stem cell-derived cardiomyocytes in 2D monolayer culture vs. 3D engineered heart tissue. Cardiovasc. Res. 2021, 117, 2125–2136.
- Kim, B.-C.; Kwack, K.; Chun, J.; Lee, J.-H. Comparative transcriptome analysis of human adipose-derived stem cells undergoing osteogenesis in 2D and 3D culture conditions. Int. J. Mol. Sci. 2021, 22, 7939.
- Nassor, F.; Jarray, R.; Biard, D.S.F.; Maïza, A.; Papy-Garcia, D.; Pavoni, S.; Deslys, J.-P.; Yates, F. Long Term Gene Expression in Human Induced Pluripotent Stem Cells and Cerebral Organoids to Model a Neurodegenerative Disease. Front. Cell. Neurosci. 2020, 14, 14.
- Yanagi, T.; Kajiya, H.; Fujisaki, S.; Maeshiba, M.; Yanagi, S.A.; Yamamoto-M, N.; Kakura, K.; Kido, H.; Ohno, J. Three-dimensional spheroids of dedifferentiated fat cells enhance bone regeneration. Regen. Ther. 2021, 18, 472–479.
- Zhu, Y.; Kang, E.; Wilson, M.; Basso, T.; Chen, E.; Yu, Y.; Li, Y.-R. 3D Tumor Spheroid and Organoid to Model Tumor Microenvironment for Cancer Immunotherapy. Organoids 2022, 1, 12.
- Tuveson, D.A.; Clevers, H. Cancer modeling meets human organoid technology. Science 2019, 364, 952–955.
- Hofer, M.; Lutolf, M.P. Engineering organoids. Nat. Rev. Mater. 2021, 6, 402–420.
- Parihar, A.; Pandita, V.; Khan, R. 3D printed human organoids: High throughput system for drug screening and testing in current COVID-19 pandemic. Biotechnol. Bioeng. 2022, 119, 2669–2688.
- Shabalina, E.Y.; Skorova, E.Y.; Chudakova, D.A.; Anikin, V.B.; Reshetov, I.V.; Mynbaev, O.A.; Petersen, E.V. The matrix-dependent 3D spheroid model of the migration of non-small cell lung cancer: A step towards a rapid automated screening. Front. Pharmacol. 2021, 8, 115.
- Nath, S.; Devi, G.R. Three-Dimensional Culture Systems in Cancer Research: Focus on Tumor Spheroid Model. Pharmacol. Ther. 2016, 163, 94–108.
- Filipiak-Duliban, A.; Brodaczewska, K.; Kajdasz, A.; Kieda, C. Spheroid Culture Differentially Affects Cancer Cell Sensitivity to Drugs in Melanoma and RCC Models. Int. J. Mol. Sci. 2022, 23, 1166.
- Liao, W.; Wang, J.; Xu, J.; You, F.; Pan, M.; Xu, X.; Weng, J.; Han, X.; Li, S.; Li, Y.; et al. High-throughput three-dimensional spheroid tumor model using a novel stamp-like tool. J. Tissue Eng. 2019, 10, 2041731419889184.
- Wang, P.; Sun, Y.; Shi, X.; Shen, H.; Ning, H.; Liu, H. 3D printing of tissue engineering scaffolds: A focus on vascular regeneration. Bio-Des. Manuf. 2021, 4, 344–378.
- Shabbirahmed, A.M.; Sekar, R.; Gomez, L.A.; Sekhar, M.R.; Hiruthyaswamy, S.P.; Basavegowda, N.; Somu, P. Recent Developments of Silk-Based Scaffolds for Tissue Engineering and Regenerative Medicine Applications: A Special Focus on the Advancement of 3D Printing. Biomimetics 2023, 8, 16.
- Chowdhury, S.R.; Lokanathan, Y.; Xian, L.J.; Busra, F.M.; Yazid, M.D.; Sulaiman, N.; Lahiry, G.; Hoque, E. 3D Printed Bioscaffolds for Developing Tissue-Engineered Constructs. In Design and Manufacturing; IntechOpen: London, UK, 2020.
- Chioni, A.-M.; Bajwa, R.T.; Grose, R. 3D Organotypic Culture Model to Study Components of ERK Signaling. Methods Mol. Biol. 2017, 1487, 255–267.
- De Gregorio, V.; La Rocca, A.; Urciuolo, F.; Annunziata, C.; Tornesello, M.L.; Buonaguro, F.M.; Netti, P.A.; Imparato, G. Modeling the epithelial-mesenchymal transition process in a 3D organotypic cervical neoplasia. Acta Biomater. 2020, 116, 209–222.
- Vernazza, S.; Tirendi, S.; Scarfì, S.; Passalacqua, M.; Oddone, F.; Traverso, C.E.; Rizzato, I.; Bassi, A.M.; Saccà, S. 2D- and 3D-cultures of human trabecular meshwork cells: A preliminary assessment of an in vitro model for glaucoma study. PLoS ONE 2019, 14, e0221942.
- Muguruma, K.; Nishiyama, A.; Kawakami, H.; Hashimoto, K.; Sasai, Y. Self-Organization of Polarized Cerebellar Tissue in 3D Culture of Human Pluripotent Stem Cells. Cell Rep. 2015, 10, 537–550.
- Kim, Y.H.; Choi, S.H.; D’Avanzo, C.; Hebisch, M.; Sliwinski, C.; Bylykbashi, E.; Washicosky, K.J.; Klee, J.B.; Brüstle, O.; Tanzi, R.E.; et al. A 3D human neural cell culture system for modeling Alzheimer’s disease. Nat. Protoc. 2015, 10, 985–1006.
- Choi, S.H.; Kim, Y.H.; Quinti, L.; Tanzi, R.E.; Kim, D.Y. 3D culture models of Alzheimer’s disease: A road map to a ‘cure-in-a-dish’. Mol. Neurodegener. 2016, 11, 75.
- Chen, H.J.; Miller, P.; Shuler, M.L. A pumpless body-on-a-chip model using a primary culture of human intestinal cells and a 3D culture of liver cells. Lab Chip 2018, 18, 2036–2046.
- Lv, D.; Hu, Z.; Lu, L.; Lu, H.; Xu, X. Three-dimensional cell culture: A powerful tool in tumor research and drug discovery (Review). Oncol. Lett. 2017, 14, 6999–7010.
- Danielson, J.J.; Perez, N.; Romano, J.D.; Coppens, I. Modelling Toxoplasma gondii infection in a 3D cell culture system In Vitro: Comparison with infection in 2D cell monolayers. PLoS ONE 2018, 13, e0208558.
- Park, S.E.; Georgescu, A.; Huh, D. Organoids-on-a-chip. Science 2019, 364, 960–965.
- Li, X.; Ootani, A.; Kuo, C. An air–liquid interface culture system for 3D organoid culture of diverse primary gastrointestinal tissues. Methods Mol. Biol. 2016, 1422, 33–40.
- Esser, L.K.; Branchi, V.; Leonardelli, S.; Pelusi, N.; Simon, A.G.; Klümper, N.; Ellinger, J.; Hauser, S.; Gonzalez-Carmona, M.A.; Ritter, M.; et al. Cultivation of Clear Cell Renal Cell Carcinoma Patient-Derived Organoids in an Air-Liquid Interface System as a Tool for Studying Individualized Therapy. Front. Oncol. 2020, 10, 1775.
- Giandomenico, S.L.; Mierau, S.B.; Gibbons, G.M.; Wenger, L.M.D.; Masullo, L.; Sit, T.; Sutcliffe, M.; Boulanger, J.; Tripodi, M.; Derivery, E.; et al. Cerebral organoids at the air–liquid interface generate diverse nerve tracts with functional output. Nat. Neurosci. 2019, 22, 669–679.
- Lin, Y.; Jiang, L.; He, Q.; Yuan, M.; Cao, J. Progress and perspective of organoid technology in cancer-related translational medicine. Biomed. Pharmacother. 2022, 149, 112869.
- Turhan, A.G.; Hwang, J.W.; Chaker, D.; Tasteyre, A.; Latsis, T.; Griscelli, F.; Desterke, C.; Bennaceur-Griscelli, A. iPSC-Derived Organoids as Therapeutic Models in Regenerative Medicine and Oncology. Front. Med. 2021, 8, 1838.
- Luo, C.; Lancaster, M.A.; Castanon, R.; Nery, J.R.; Knoblich, J.A.; Ecker, J.R. Cerebral Organoids Recapitulate Epigenomic Signatures of the Human Fetal Brain. Cell Rep. 2016, 17, 3369–3384.
- Sun, L.; Hui, L. Progress in human liver organoids. J. Mol. Cell Biol. 2020, 12, 607–617.
- Balak, J.R.A.; Juksar, J.; Carlotti, F.; Nigro, A.L.; de Koning, E.J.P. Organoids from the Human Fetal and Adult Pancreas. Curr. Diabetes Rep. 2019, 19, 160.
- Puschhof, J.; Pleguezuelos-Manzano, C.; Clevers, H. Organoids and organs-on-chips: Insights into human gut-microbe interactions. Cell Host Microbe 2021, 29, 867–878.
- Manafi, N.; Shokri, F.; Achberger, K.; Hirayama, M.; Mohammadi, M.H.; Noorizadeh, F.; Hong, J.; Liebau, S.; Tsuji, T.; Quinn, P.M.; et al. Organoids and organ chips in ophthalmology. Ocul. Surf. 2020, 19, 1–15.
- Hsia, G.S.P.; Esposito, J.; da Rocha, L.A.; Ramos, S.L.G.; Okamoto, O.K. Clinical Application of Human Induced Pluripotent Stem Cell-Derived Organoids as an Alternative to Organ Transplantation. Stem Cells Int. 2021, 2021, 6632160.
- Sugimoto, S.; Kobayashi, E.; Fujii, M.; Ohta, Y.; Arai, K.; Matano, M.; Ishikawa, K.; Miyamoto, K.; Toshimitsu, K.; Takahashi, S.; et al. An organoid-based organ-repurposing approach to treat short bowel syndrome. Nature 2021, 592, 99–104.
- Zhao, Z.; Chen, X.; Dowbaj, A.M.; Sljukic, A.; Bratlie, K.; Lin, L.; Fong, E.L.S.; Balachander, G.M.; Chen, Z.; Soragni, A.; et al. Organoids. Nat. Rev. Methods Prim. 2022, 2, 94.
