Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Cell Biology
Human dental pulp stem cells (hDPSCs) are adult mesenchymal stem cells (MSCs) obtained from dental pulp and derived from the neural crest. They can differentiate into odontoblasts, osteoblasts, chondrocytes, adipocytes and nerve cells, and they play a role in tissue repair and regeneration.
- hDPSCs
- PIEZO1
- ATP
- migracytosis
- blebbing
1. Introduction
Adult stem cells are a resource for living organisms that allow for the repair and/or regeneration of damaged tissues. In general, there are two different approaches to regenerate damaged tissue using stem cells: cell homing and cell transplantation, both of which imply cell migration. Mesenchymal stem cells (MSCs) are found mainly in the bone marrow (BM), but also in the adipose tissue and in the pulp of the tooth. They can give rise to osteoblasts, chondrocytes and adipocytes. Dental pulp stem cells (DPSCs) are unique because they arise from the ectomesodermal embryonic tissue that forms the neural crest. For this reason, in addition to the cell types described above, they can give rise to odontoblasts (specialized osteoblasts) and nerve cells (astrocytes, glia cells and oligodendrocytes). The microenvironment in which stem cells are found affects their differentiation. In the case of naïve DPSCs, the niches in which they are located are innervated, supplied with blood and inside a rigid structure (tooth). The decision between the renewal and migration/differentiation of DPSCs depends on their interactions with stromal cells and ECMs in the niches. Different niches possess different DPSCs. As an example, after tooth injury in the apical part of the pulp, there was a population of highly proliferative potential which was Notch2-positive [1], whereas in the perivascular niches, DPSCs were positive for Oct3/4 stemness markers [2]. It has been shown that in vivo human (h) DPSCs migrate and can repair dentin by regenerating damaged odontoblasts in the tooth [3]. Furthermore, hDPSCs transplanted into mice can promote bone regeneration in defective calvaria [4] or migrate to ischemic areas, i.e., areas of cerebral infarction, and express specific neural markers [5]. Additionally, when induced in vitro as neural cells, they can differentiate in vivo into mature neurons or astrocytes [6]. However, hDPSCs can also repair damaged nerve tissue through paracrine mechanisms involving chemotaxis and the proliferation of endogenous neural stem cells (NSCs) [7], or they can reduce ischemic damage through the inhibition of microglial activation and the expression of pro-inflammatory cytokines [8]. When comparing MSC transplantation with the practice of homing for tissue regeneration in preclinical animal models, the latter is safer and more effective [9].
2. HDPSCs
HDPSCs can be purified from dental pulp essentially by two methods. The first is based on the enzymatic digestion of the dental pulp with collagenase and dispase and/or trypsin [10] (Table 1).
Table 1. Schematic list of the manuscripts cited divided according to the DPSC recovery method.
The harvested cells were dispersed in the medium, plated and left to proliferate. In turn, they formed colonies of heterogeneous cells, including stem cells from various niches in a mixture, and epithelial cells, stromal cells, perivascular cells, etc. [19] (Figure 1A). In this regard, heterogeneous cells expanded in a serum-free medium produced two DPSC subtypes, those being adherent (ADH) and non-adherent (non-ADH) populations according to their differential adhesion to plastic; however, both populations displayed osteogenic and neurogenic differentiation [26,48]. The second method consisted of putting the pulp, or fragments, directly in a plate with a culture medium and waiting for the DPSCs to come out after about 10–15 days (explant method) (Table 1, Figure 1B and Figure 2A) [16]. The tissue piece is present during the primary culture and therefore, DPSCs reside in the dental niches with stromal cells and extracellular matrix (ECM), since no proteolytic enzymes are added in the culture. DPSCs take a long time to come out from the pulp, but they take advantage of the presence of other cells and the ECM. This method makes it possible to observe the cells that are induced to migrate. Indeed, DPSCs gradually emerge from the tissue, whereas non-migrating cells remain inside the tissue and can migrate later (for agreement with DPSCs residing in different niches, see [1], or if they are not stem cells, they undergo apoptosis [1,2,49]). However, DPSCs obtained by the explant method produce a more homogeneous population (i.e., subsequent waves, see [20]), and unattached and adherent cells, different from DPSCs, are present in the culture [24]. However, unattached cells will be gradually removed after refreshing the culture media, and adherent cells are unable to survive/proliferate and will be lost during the first few subcultures [20,49].
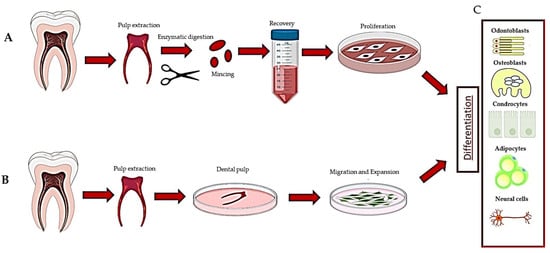
Figure 1. Description of the methods used to recover dental pulp stem cells: (A) digestion method, (B) explant method and (C) differentiation of DPSCs.
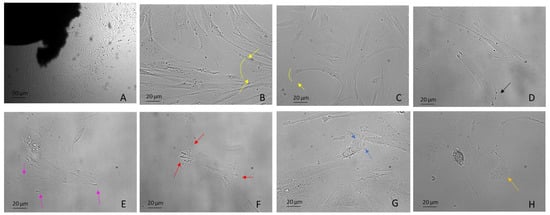
Figure 2. DPSCs harvested using the explant method from the third molar extracted for orthodontic reasons. Examples include: (A) DPSCs recovered by explant method (10×) (B,C) mesenchymal migration (20×), (D) FA (20×), (E) lamellipodia (20×), (F) filopodia (20×), (G) blebs and ameboid migration (20×), (H) migrasomes (20×). Light Microscopy (Zeiss Axio). DPSCs were isolated with the explant method from teeth removed for orthodontic reasons and the patients agreed to the use of their samples for research purposes. Bar = 20 µm; 50 µm.
Therefore, the two isolation methods yielded different subpopulations of cells, even though regardless of the recovery method, DPSCs showed the same trilineage differentiation potential when not pre-selecting for specific marker expressions [24,27,50]. In general, in vitro hDPSCs are most likely induced to migrate and proliferate following environmental cues such as chemical and biophysical stimuli, for example, changes in stiffness and rigidity (both are mechanical stimuli) [51,52]. However, in the explant method, a wound healing response is triggered due to the production of cytokines and factors released by the injured tissue, which promote migration [30,49]. Cells harvested with the explant method migrate as “leaders” and “followers”, where leaders migrate first as single cells [28,53] and guide the migration and followers follow the guide, connoting a subdivision of the group into distinct fractions [54].
Many mathematical models (stochastic models) have been developed to explain the various types of cell migration. However, these models are often based on the migration of clustered cells (tumors) or activated lymphocytes (taxis), whereas DPSCs, as said before, can migrate as single cells [55]. In addition, some models are based on the idea that the nucleus occupies a central position, but this is not always true. For example, in DPSCs, the nucleus is often lateral or in the back. Moreover, in response to environmental cues, many cells have the capacity to turn off their default migration mode from mesenchymal to ameboid and vice versa [56]. Another important feature to take into consideration is given by the stimuli that recruit the “leaders”, which is not fully understood and can be single or double. To date, the literature is still scarce concerning mathematical models that explain cell migration in the presence of two stimuli. The first cue concerns the choice of direction and the second, usually of mechanical origin, concerns the speed that the cell can reach going in that direction [57,58,59]. Speed is important because the fastest cells (leader) can lead the others (follower). However, further studies are needed to define a good mathematical model that accounts for the migration of DPSCs.
This entry is adapted from the peer-reviewed paper 10.3390/biology12050742
This entry is offline, you can click here to edit this entry!
