Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Esophageal cancer (EC) is the deadliest cancer worldwide, with a 92% annual mortality rate per incidence. Esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC) are the two major types of ECs, with EAC having one of the worst prognoses in oncology. Limited screening techniques and a lack of molecular analysis of diseased tissues have led to late-stage presentation and very low survival durations.
- esophageal carcinoma
- esophageal adenocarcinoma
- biomarkers
1. Introduction
Its poor prognosis and high mortality rate make esophageal cancer (EC) one of the deadliest cancers worldwide. EC is the eighth most common cancer worldwide (the seventh most common cancer in men and the thirteenth most common cancer in women) and the sixth leading cause of cancer-related deaths. The two most common histological types of EC, namely esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC), account for more than 90% of ECs. The five-year survival rate of EC is less than 20% [1][2][3][4]. A recent retrospective study reporting 23,804 EAC cases and 13,919 ESCC cases suggests an increasing incidence of EAC and a decreasing incidence of ESCC in the United States [3]. ESCC is characterized by the conversion of the normal squamous esophageal epithelium to ESCC via basal cell hyperplasia, dysplasia, and invasiveness. ESCC may involve any part of the esophagus (20% upper, 50% middle, and 30% lower esophagus). Alcohol and tobacco consumption, the most common risk factors, cause cellular DNA damage and contribute to ESCC. EAC occurring in the distal esophagus occurs due to a cascade progressing from gastroesophageal reflux disease (GERD) to Barrett’s esophagus (BE), followed by EAC with columnar metaplasia playing a critical role in the pathogenesis (Figure 1). Male sex, white race, central obesity, alcohol, and smoking are common risk factors, while inflammation, genetic mutations, epigenetics, and altered microbiota play a critical role in the pathogenesis of EAC [4][5][6][7][8].
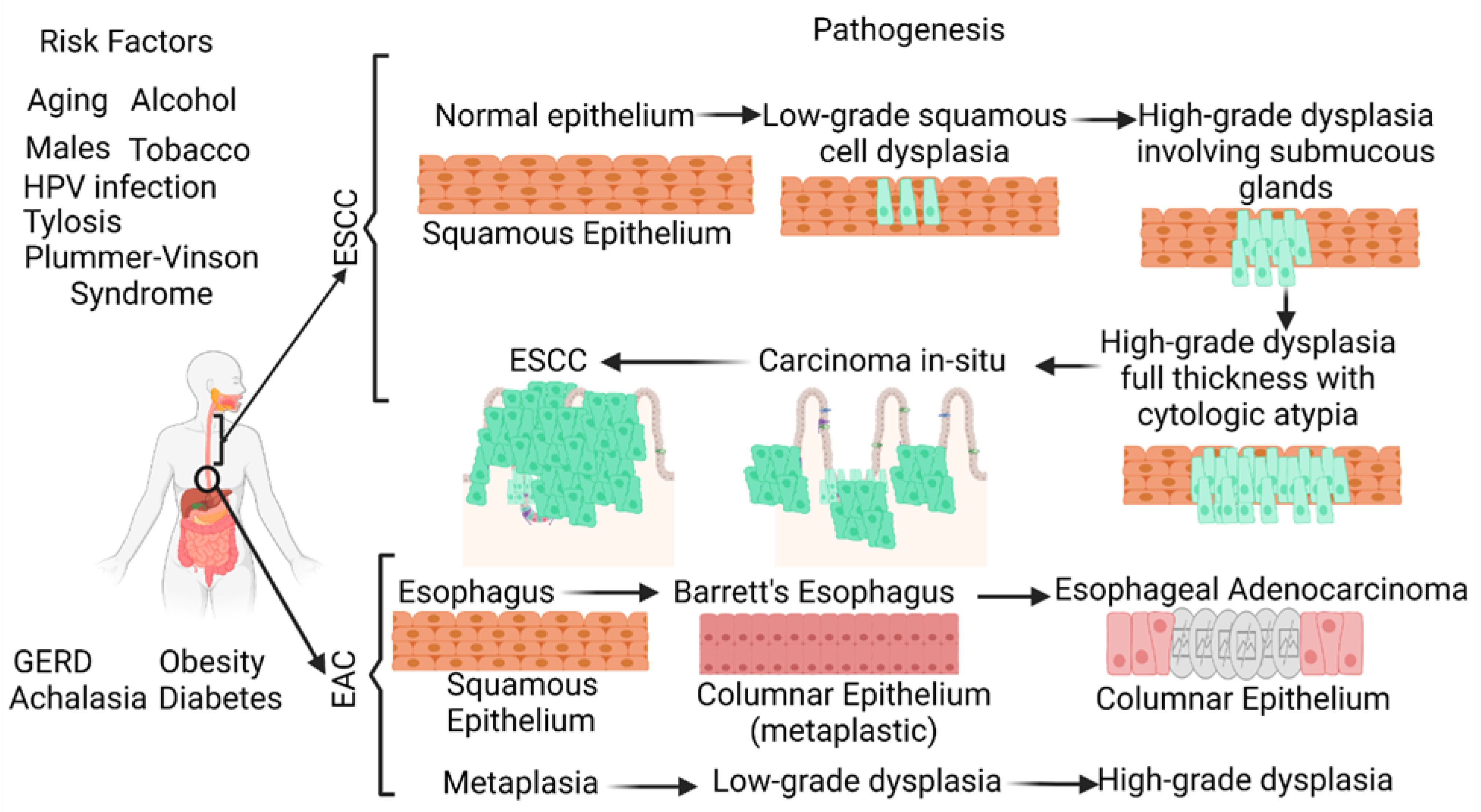
Figure 1. Schematic diagram showing the risk factors and pathogenesis of esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC). ESCC develops through a multistep process from basal hyperplasia due to chronic esophagitis through increasing severity of dysplasia, while EAC develops through a cellular cascade involving gastroesophageal reflux disease (GERD) followed by Barrett’s esophagus leading to EAC. There are different risk factors in the pathogenesis of ESCC and EAC. HPV, human papillomavirus.
Early detection of ESCC and EAC is needed to improve survival and attenuate morbidity and mortality. Esophageal endoscopy with biopsy and histological analysis is the gold standard for early detection and diagnosis. Chromoendoscopy, virtual chromoendoscopy, magnification endoscopy, and other advanced endoscopic imaging techniques may improve the sensitivity of the detection of early-stage carcinoma [9]. However, difficulty in defining a well-characterized screening population, the lack of an accurate, cost-effective, and widely accepted screening tool, and the absence of data on the costs of non-invasive screening are associated challenges [10]. Additionally, endoscopic screening is not practical for mass screening because of the invasive and expensive procedures involved. Surgical resection is the most common treatment for early-stage EC, and chemotherapy, radiotherapy, chemoradiation, laser therapy, electrocoagulation, immunotherapy, and targeted therapy are treatment strategies for advanced and nonresectable lesions [11]. The current treatment regimen for EAC is based on the expression of a number of biomarkers, including human epidermal growth factor receptor 2 (HER2) amplification, mismatch repair deficiency/microsatellite instability (dMMR/MSI-H), and programmed death ligand 1 (PD-L1) [12]. Early-stage disease and complete resection of the lesion are favorable prognostic markers [13]. However, the ineffectiveness of chemotherapy, radiation therapy, immunotherapy, and targeted therapy contributing to low survival rates warrants the establishment of early-stage diagnostic biomarkers and novel therapeutics to improve clinical outcomes and decrease morbidity and mortality [14][15]. This notion is further supported by the asymptomatic nature of EC in its early stages, its extremely aggressive nature, and its poor survival rate.
2. Biomarkers for EC: Pros and Cons
Poor prognosis due to late detection of ECs warrants the development of early detection methods using non-invasive biomarkers so that timely intervention can be started to improve outcomes. Tissue histology after endoscopy has limitations for mass screening, and serum tumor markers including squamous cell carcinoma antigen (SCCA) and carcinoembryonic antigen (CEA) are insufficiently specific and sensitive for early EC diagnosis [16][17]. Lesion recognition during endoscopy is impeded by inter- and intra-observer variability [18]. Blood biomarkers/liquid biopsy (circulating tumor cells, nucleic acids, tumor DNA, the tumor-derived fraction of cell-free DNA, cell-free RNA, etc.) has higher specificity and accuracy. Proteomic profiling has potential, but is limited due to higher costs in routine use, and epigenetic markers are promising due to ease of detection in tissue and body fluids including blood, plasma, and urine. Liquid biopsy is advantageous in the case of metastatic tumors which are difficult sample using a core biopsy [19]. RNA biomarkers (including mRNA, miRNA, and long non-coding RNA), protein biomarkers, metabolic biomarkers, immune biomarkers, and microbiome biomarkers are commonly documented biomarkers for EC diagnosis [20][21][22]. In addition to tissue-based or liquid biopsy-based biomarkers, imaging-based biomarkers including perfusion analysis using computed tomography (CT) or magnetic resonance imaging (MRI), texture analysis, diffusion-weighted imaging (DWI), and positron emission tomography (PET) may also be used in the treatment of EC using radiomics, an emerging field in which imaging data are converted into a high dimensional mineable feature [23]. Imaging biomarkers may have the potential to predict treatment outcomes or prognosis in EC due to their non- or less-invasive nature and wider availability.
3. Non-Invasive Biomarkers: Blood, Plasma, Saliva, and Urine Biomarkers
Liquid biopsy and blood biomarkers are gaining attention because of their non-invasive nature, simplicity, short-term repeatability, and cost-effectiveness, as well as their ability to detect circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and exosome-based biomarkers for both EAC and ESCC [24]. Urinary protein may serve as a biomarker for ESCC. Li et al. [25] conducted a proteomics analysis on 499 human urine samples (321 healthy individuals, 83 with ESCC, 17 with bladder cancer, 12 with breast cancer, 16 with colorectal cancer, 33 with lung cancer, and with 17 thyroid cancer). The results suggested that urinary biomarkers ANXA1, S100A8, and TMEM256 can classify ESCC, and a panel of proteins consisting of ANXA1, S100A8, SOD3, and TMEM256 is diagnostic for stage I ESCC. Further, serum expression of other factors involved in the pathogenesis of EAC and ESCC may also be potential biomarkers, e.g., chemokines and chemokine receptors. CXCL12 and its receptors CXCR4 and CXCR7 correlate with poor prognosis, CXCL10, CCL4, and CCL5 expression show anti-tumor effect, CCL20 expression is correlated with regulatory T cell recruitment, and CCR7 expression correlates with poor prognosis [26]. Additionally, neutrophil-lymphocyte ratio (NLR) [27], erythrocyte mutant frequency (EMF) [28], and serum antibodies (anti-p53, anti-p16, anti-cyclin B1, anti-c-Myc, anti-HSP70, and anti-LY6K) may potentially play a role [18] in the diagnosis of EAC and its differentiation from BE. Using saliva as a non-invasive sample for biomarkers is useful not only for oral cancers but also for non-oral cancers [29]. Liquid biopsy and blood biomarkers offer an inexpensive and non-invasive screening strategy and the use of advanced technologies, such as metabolomics and proteomics in combination, has allowed the delineation of novel diagnostic biomarkers for the early detection of ESCC and EAC [18]. However, using a single serum marker for early detection and diagnosis may have low diagnostic value, and using a panel of biomarkers in combination can significantly improve the sensitivity and specificity of the early detection and diagnosis of ESCC and EAC.
Taken together, the detection of a panel of non-invasive biomarkers in blood, urine, and saliva may increase diagnostic sensitivity and specificity and have a potential clinical application in improving outcomes [30]. Using non-invasive biomarkers in clinics will be useful because analysis of the non-invasive biomarkers utilizes readily available clinical and laboratory information to non-invasively detect the tumor early in course of disease in at-risk populations and can be applied to mass screening. Other advantages of using non-invasive strategies are the absence of adverse effects and the attenuated risk of sampling error. This will bring objectiveness to the interpretation and can overcome the limitations of endoscopy for mass screening. Further, non-invasive biomarkers are not only useful in early diagnosis, but they also play a role in predicting the treatment outcome, disease progression, and relapse [31][32]. Although the non-invasive biomarkers from saliva and urine that can be used in clinics for early detection of ESCC and EAC are limited, the results from various studies, outlined in Table 1, suggest that in addition to liquid biopsy, non-invasive samples such as urine and saliva may be used for detecting biomarkers in both ESCC and EAC.
Table 1. Non-invasive biomarkers for esophageal carcinoma. Esophageal squamous cell carcinoma (ESCC), esophageal adenocarcinoma (EAC), gelsolin (GSN), serum paraoxonase/arylesterase 1 (PON1) and serum paraoxonase/lactonase 3 (PON3), desmoglein-2 (DSG2), serum amyloid A1 (SAA1), enolase 1 (ENO1), triosephosphate isomerase 1 (TPI1), toll-like receptor (TLR)-4, hypoxia-inducible factor (HIF)-1α, tricarboxylic acid (TCA) cycle, deoxynivalenol (DON), neosolaniol (NEO), T-2 toxin (T-2), HT-2 toxin (HT-2).
| Sample Type | EAC/ESCC | Sample Size | Biomarker Type/Observation |
|---|---|---|---|
| Serum [33] | EAC | 159 EAC patients | Metabolomic profiling; among D-mannose, L-proline (LP), and 3-hydroxybutyrate (BHBA) were significantly different in the EAC patients and in the controls; the serum level of D-mannose may be a novel prognostic biomarker for EAC |
| Serum [34] | EAC | 301 samples | To identify glycoprotein biomarkers; different glycoforms of complement C9 (C9), GSN, PON1, and PON3 are biomarkers for EAC and discriminate it from BE; serum levels of C9 glycoforms increase with disease progression |
| Saliva [35] | EAC | DNA methylation profiles for 125 EAC and 64 normal adjacent squamous samples; saliva samples from 192 patients |
A proto-cadherin module centered around CTNND2 is inactivated in Barrett’s esophagus; CCL20 chemokine methylation pattern in saliva correlates with EAC status |
| Serum [36] | ESCC, EJA | 151 ESCC and 96 EJA cases with 212 healthy controls. | Serum DSG2 was significantly higher in ESCC and EJA compared with controls; serum DSG2 levels were significantly associated with patient age and histological grade in ESCC; serum DSG2 may be a biomarker for ESCC and EJA |
| Serum [37] | ESCC | 30 ESCC patients and 30 healthy controls | Serum proteins S100A8/A9, SAA1, ENO1, TPI1, and PGAM1 have high diagnostic sensitivity and specificity for ESCC; glycolysis, TLR4, HIF-1α, Cori cycle, TCA cycle, folate metabolism, and platelet degranulation are commonly deregulated pathways |
| Saliva [38] | ESCC | 178 ESCC patients and 101 healthy controls | Significantly higher numbers of Streptococcus salivarius, Fusobacterium nucleatum, and Porphyromonas gingivalis in patients with ESCC suggest salivary microbiota as a biomarker |
| Urine [25] | ESCC | 499 urine samples (83 ESCC) | ANXA1, S100A8, and TMEM256 can classify ESCC; a combination panel of the proteins ANXA1, S100A8, SOD3, and TMEM256 is diagnostic for stage I ESCC |
| Serum [39] | EC | 20 EC patients and 20 healthy controls | Serum anaphylatoxin C3a may be a promising biomarker in the diagnosis of EC |
| Urine [40] | EC | 10 controls and 17 EC patients | Mycotoxins as binary (NEO/HT-2 and T-2/HT-2) and ternary (DON/NEO/HT-2) combinations were present in the urine samples of patients with EC |
4. Molecular Biomarkers
microRNA (miR), genetically conserved small noncoding RNA of 18–25 nucleotides, regulates gene expression by binding to the 3′-UTR of target mRNAs, post-transcriptionally resulting in either translational inhibition or degradation of RNA. This will cause the gene expression to be either upregulated or downregulated; activation of miRs downregulates gene expression while decreased miR expression upregulates gene expression [41][42]. The involvement of miRs in EC tumorigenesis and progression and their identification as a biomarker in blood, plasma, and urine suggests that miRs may be a potential non-invasive biomarker [41]. Fassan et al. [43] reported upregulation of miR-92a-3p, miR-151a-5p, miR-362-3p, miR-345-3p, miR-619-3p, miR-1260b, and miR-1276 as well as downregulation of miR-381-3p, miR-502-3p, and miR-3615 in the serum of early EAC patients compared with non-dysplastic BE. Further, Chiam et al. [44] reported that the ratios of RNU6-1/miR-16-5p, miR-25-3p/miR-320a, let-7e-5p/miR-15b-5p, miR-30a-5p/miR-324-5p, and miR-17-5p/miR-194-5p in circulating exosomes with an AUC of 0.99 could differentiate between EAC and nondysplastic BE. When looking for small extracellular vesicle microRNAs as biomarkers for EAC, the serum is more suitable than the plasma [45]. Thus, miRs are useful as biomarkers for diagnosis and, before surgery, to predict chemotherapy outcomes [46].
Circulating tumor DNA (ctDNA), the DNA coming out from cancerous cells and tumors and circulating in the blood, may be a potential biomarker for early diagnosis. However, detection of ctDNA in the early stages of EAC is challenging and may have limited diagnostic application [47]. Further, ctDNA levels and the detection of its variants were also found to be associated with poor survival, and the variant frequency increased with recurrence [48]. The potential of ctDNA as a biomarker and its use to monitor improvement and relapse was supported by the detection of a suitable number of somatic single-nucleotide variants (SNVs) and copy number alterations (CNAs) in the plasma of EAC patients using sequencing and a NanoString Counter [49]. Further, the detection of post-operative ctDNA provides a molecular window before the onset of overt disease and allows researchers to add another therapy to improve outcomes. These studies suggest the potential clinical utility of ctDNA as a prognostic biomarker for early diagnosis, monitoring treatment response and disease recurrence, and improving survival with moderate sensitivity and high specificity. This ability is further enhanced when combined with current imaging methods [50]. Additionally, cell-free plasma DNA and exosome-associated DNA from blood [19] may also be used as a biomarker for EAC diagnosis. Circulating cell-free DNA has diagnostic value and targets tumor-specific genomes by detecting epigenetic (methylation of APC, CDKN2A, TAC1, and MSH2) and genetic alterations that might have translational and clinical significance and may be more reliable than the existing biomarkers such as CEA [51]. Additionally, circular RNAs (circRNAs), which play a role in cell proliferation, migration, death, tumor invasion, and metastasis, may also be used as biomarkers for ESCC because dysregulated expression of circRNA is associated with the pathogenesis of ESCC and it can be detected not only in tumor tissue but also in nearby tissue. The detection of circRNAs using techniques such as RNA sequencing and bioinformatics analysis enables the detection of both known and unknown circRNAs, which is beneficial compared with the microarray technique, which detects only known circRNA [52]. In addition to circRNA, miRNAs, and ctDNAs, transcription factors (TFs), the regulators of gene expression, may also serve as biomarkers for early detection. TFs, including BRCA1, SOX10, ARID3A, ZNF354C, and NFIC, play a role in carcinogenesis and the development of ESCC, while SREBF1 and TFAP2A correlated with longer overall survival in ESCC. These TFs may also serve as diagnostic biomarkers [53]. Various studies [30][54] and reports, summarized in Table 2, indicate the role of microRNAs, tRNA-derived small RNAs, circulating tumor (ct) DNAs, and transcription factors as biomarkers in esophageal carcinoma.
Table 2. Molecular biomarkers for ESCC and EAC. Area under the curve (AUC), esophageal adenocarcinoma (EAC), esophageal squamous cell carcinoma (ESCC), healthy control (HC), circulating tumor (ct) DNA, Barrett’s esophagus (BE), next-generation sequencing (NGS), variants of unknown significance (VUS), gelsolin (GSN), serum paraoxonase/arylesterase 1 (PON1) and serum paraoxonase/lactonase 3 (PON3), esophageal squamous cell carcinoma (ESCC) and esophagogastric junction adenocarcinoma (EJA), desmoglein-2 (DSG2), tRNA-derived small RNAs (tsRNAs).
| Sample Type | EAC/ESCC | Sample Size | Biomarker Type/Observation |
|---|---|---|---|
| Urine [41] | EAC and ESCC | 150 HCs and 43 ESCCs 144 HCs and 8 EACs |
Significantly higher miR-1273f, miR-619-5p, miR-150-3p, miR-4327, and miR-3135b levels in ESCC and EAC compared with HCs; miR-1273f and miR-619-5p with AUC ≥ 0.80 for diagnosing stage I ESCC, AUC ≥ 0.80 in ESCC, and AUC = 0.80 for EAC |
| Urine, saliva, and blood [55] | ESCC | 72 ESCC patients | Serum cell-free miR-1246 expression in the urine, saliva, and serum may be a useful biomarker for ESCC and urine can be used as a non-invasive sample instead of blood |
| Plasma [56] | ESCC | 16 healthy controls and 66 ESCC patients | Plasma miR-21, miR-31, and miR-375 could be potential biomarkers for the diagnosis of ESCC, while miR-31 and miR-375 have sufficiently high sensitivity and specificity to differentiate ESCC patients from healthy controls |
| Saliva [57] | EC | miRNA expression profile GSE41268 | miR-144, miR-451, miR-98, miR-10b, and miR-363 may serve as biomarkers for EC |
| Saliva [58] | 32 EC patients and 16 healthy controls | Salivary supernatant miR-21 was significantly higher in EC with a sensitivity and specificity of 84.4% and 62.5%, respectively; miR-21 expression does not correlate with EC stage | |
| Saliva [59] | EC | 7 EC patients and 3 healthy controls | miR-10b*, miR-144, and miR-451 in the whole saliva and miR-10b*, miR-144, miR-21, and miR-451 in saliva supernatant were significantly upregulated in patients, with sensitivity and specificity ranging between 43.6% and 92.3% |
| Saliva [60] | ESCC | 3 ESCC patients and 3 healthy controls |
RNA sequencing of salivary exosomes for identification of tsRNA; tsRNA (tRNA-GlyGCC-5) was significantly enriched in salivary exosomes in ESCC |
| Plasma [47] | EAC | Patients with stage I to IV EAC 55 tumor and matched normal samples |
Detection frequency and quantity of ctDNA increase with stage; ctDNA positively correlates with disease burden; ctDNA levels during the treatment may be useful to determine response and recurrence in some patient |
| Plasma [48] | EAC | 209 blood and tumor samples from 57 EAC patients | Both plasma and tumor samples were sequenced for ctDNA; detectable ctDNA variants in post-treatment plasma samples were associated with worse disease-specific survival; variant allele frequency of ctDNA variants increased with disease recurrence |
| Plasma [61] | BE EAC |
138 patients: EAC = 41 Barrett’s dysplasia = 48 Control = 49 |
To detect circulating HPV DNA; higher circulating HPV DNA was detected in EAC patients with invasive tumors with submucosal invasion and lymph node metastasis; circulating HPV DNA positivity was associated with tissue HPV positivity and disease severity |
| Plasma [49] | EAC | 40 EAC patients (17 palliative and 23 curative) | Sensitive ctDNA detection has potential for the monitoring and predicting of short overall survival; the presence of ctDNA post-operatively predicts relapse and provides a molecular window before the onset of overt disease |
| Plasma [62] | EAC | 55 EAC patients with advanced disease | ctDNA detection using NGS; 66% of patients had ≥ 1 genomic alteration including VUS and 69.1% had ≥ 1 characterized alteration (excluding VUSs); patients with ≥ 1 characterized alteration had alterations targetable by an FDA-approved therapy theoretically |
This entry is adapted from the peer-reviewed paper 10.3390/ijms24043316
References
- Domper Arnal, M.J.; Ferrandez Arenas, A.; Lanas Arbeloa, A. Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J. Gastroenterol. 2015, 21, 7933–7943.
- Short, M.W.; Burgers, K.G.; Fry, V.T. Esophageal Cancer. Am. Fam. Physician 2017, 95, 22–28.
- Then, E.O.; Lopez, M.; Saleem, S.; Gayam, V.; Sunkara, T.; Culliford, A.; Gaduputi, V. Esophageal Cancer: An Updated Surveillance Epidemiology and End Results Database Analysis. World J. Oncol. 2020, 11, 55–64.
- Napier, K.J.; Scheerer, M.; Misra, S. Esophageal cancer: A Review of epidemiology, pathogenesis, staging workup and treatment modalities. World J. Gastrointest. Oncol. 2014, 6, 112–120.
- Watanabe, M. Risk factors and molecular mechanisms of esophageal cancer: Differences between the histologic subtypes. J. Cancer Metastasis Treat. 2015, 1, 1–7.
- Singhal, S.; Kapoor, H.; Subramanian, S.; Agrawal, D.K.; Mittal, S.K. Polymorphisms of Genes Related to Function and Metabolism of Vitamin D in Esophageal Adenocarcinoma. J Gastrointest. Cancer 2019, 50, 867–878.
- Kailasam, A.; Mittal, S.K.; Agrawal, D.K. Epigenetics in the Pathogenesis of Esophageal Adenocarcinoma. Clin. Transl. Sci. 2015, 8, 394–402.
- Gillespie, M.R.; Rai, V.; Agrawal, S.; Nandipati, K.C. The Role of Microbiota in the Pathogenesis of Esophageal Adenocarcinoma. Biology 2021, 10, 697.
- Meves, V.; Behrens, A.; Pohl, J. Diagnostics and Early Diagnosis of Esophageal Cancer. Viszeralmedizin 2015, 31, 315–318.
- Kamboj, A.K.; Katzka, D.A.; Iyer, P.G. Endoscopic Screening for Barrett’s Esophagus and Esophageal Adenocarcinoma: Rationale, Candidates, and Challenges. Gastrointest. Endosc. Clin. N. Am. 2021, 31, 27–41.
- Rubenstein, J.H.; Shaheen, N.J. Epidemiology, Diagnosis, and Management of Esophageal Adenocarcinoma. Gastroenterology 2015, 149, 302–317.e301.
- Dhakras, P.; Uboha, N.; Horner, V.; Reinig, E.; Matkowskyj, K.A. Gastrointestinal cancers: Current biomarkers in esophageal and gastric adenocarcinoma. Transl. Gastroenterol. Hepatol. 2020, 5, 55.
- Luthringer, M.; Marziale, J. Esophageal Cancer Treatment (PDQ®) Health Professional Version Last Modified: 07/13/2012. Available online: https://www.advancedob-gyn.com/health-library/hw-view.php?DOCHWID=ncicdr0000062741 (accessed on 9 January 2023).
- Abdo, J.; Wichman, C.S.; Dietz, N.E.; Ciborowski, P.; Fleegel, J.; Mittal, S.K.; Agrawal, D.K. Discovery of Novel and Clinically Relevant Markers in Formalin-Fixed Paraffin-Embedded Esophageal Cancer Specimen. Front. Oncol. 2018, 8, 157.
- Abdo, J.; Bertellotti, C.A.; Cornell, D.L.; Agrawal, D.K.; Mittal, S.K. Neoadjuvant Therapy for Esophageal Adenocarcinoma in the Community Setting-Practice and Outcomes. Front. Oncol. 2017, 7, 151.
- Nakamura, T.; Ide, H.; Eguchi, R.; Hayashi, K.; Takasaki, K.; Watanabe, S. CYFRA 21-1 as a tumor marker for squamous cell carcinoma of the esophagus. Dis. Esophagus 2017, 11, 35–39.
- Zheng, X.; Xing, S.; Liu, X.M.; Liu, W.; Liu, D.; Chi, P.D.; Chen, H.; Dai, S.Q.; Zhong, Q.; Zeng, M.S.; et al. Establishment of using serum YKL-40 and SCCA in combination for the diagnosis of patients with esophageal squamous cell carcinoma. BMC Cancer 2014, 14, 490.
- Visaggi, P.; Barberio, B.; Ghisa, M.; Ribolsi, M.; Savarino, V.; Fassan, M.; Valmasoni, M.; Marchi, S.; de Bortoli, N.; Savarino, E. Modern Diagnosis of Early Esophageal Cancer: From Blood Biomarkers to Advanced Endoscopy and Artificial Intelligence. Cancers 2021, 13, 3162.
- Smith, R.A.; Lam, A.K. Liquid Biopsy for Investigation of Cancer DNA in Esophageal Adenocarcinoma: Cell-Free Plasma DNA and Exosome-Associated DNA. Methods Mol. Biol. 2018, 1756, 187–194.
- Yang, W.; Han, Y.; Zhao, X.; Duan, L.; Zhou, W.; Wang, X.; Shi, G.; Che, Y.; Zhang, Y.; Liu, J.; et al. Advances in prognostic biomarkers for esophageal cancer. Expert Rev. Mol. Diagn. 2019, 19, 109–119.
- Li, Y.; Liu, J.; Cai, X.W.; Li, H.X.; Cheng, Y.; Dong, X.H.; Yu, W.; Fu, X.L. Biomarkers for the prediction of esophageal cancer neoadjuvant chemoradiotherapy response: A systemic review. Crit. Rev. Oncol. Hematol. 2021, 167, 103466.
- Hou, X.; Wen, J.; Ren, Z.; Zhang, G. Non-coding RNAs: New biomarkers and therapeutic targets for esophageal cancer. Oncotarget 2017, 8, 43571–43578.
- Hayano, K.; Ohira, G.; Hirata, A.; Aoyagi, T.; Imanishi, S.; Tochigi, T.; Hanaoka, T.; Shuto, K.; Matsubara, H. Imaging biomarkers for the treatment of esophageal cancer. World J. Gastroenterol. 2019, 25, 3021–3029.
- Iacob, R.; Mandea, M.; Iacob, S.; Pietrosanu, C.; Paul, D.; Hainarosie, R.; Gheorghe, C. Liquid Biopsy in Squamous Cell Carcinoma of the Esophagus and of the Head and Neck. Front. Med. 2022, 9, 827297.
- Ji, L.; Wang, J.; Yang, B.; Zhu, J.; Wang, Y.; Jiao, J.; Zhu, K.; Zhang, M.; Zhai, L.; Gong, T.; et al. Urinary protein biomarker panel predicts esophageal squamous carcinoma from control cases and other tumors. Esophagus 2022, 19, 604–616.
- Goto, M.; Liu, M. Chemokines and their receptors as biomarkers in esophageal cancer. Esophagus 2020, 17, 113–121.
- Campos, V.J.; Mazzini, G.S.; Juchem, J.F.; Gurski, R.R. Neutrophil-Lymphocyte Ratio as a Marker of Progression from Non-Dysplastic Barrett’s Esophagus to Esophageal Adenocarcinoma: A Cross-Sectional Retrospective Study. J. Gastrointest. Surg. 2020, 24, 8–18.
- Haboubi, H.N.; Lawrence, R.L.; Rees, B.; Williams, L.; Manson, J.M.; Al-Mossawi, N.; Bodger, O.; Griffiths, P.; Thornton, C.; Jenkins, G.J. Developing a blood-based gene mutation assay as a novel biomarker for oesophageal adenocarcinoma. Sci. Rep. 2019, 9, 5168.
- Rapado-Gonzalez, O.; Martinez-Reglero, C.; Salgado-Barreira, A.; Takkouche, B.; Lopez-Lopez, R.; Suarez-Cunqueiro, M.M.; Muinelo-Romay, L. Salivary biomarkers for cancer diagnosis: A meta-analysis. Ann. Med. 2020, 52, 131–144.
- Chu, L.Y.; Peng, Y.H.; Weng, X.F.; Xie, J.J.; Xu, Y.W. Blood-based biomarkers for early detection of esophageal squamous cell carcinoma. World J. Gastroenterol. 2020, 26, 1708–1725.
- Harrington, C.; Krishnan, S.; Mack, C.L.; Cravedi, P.; Assis, D.N.; Levitsky, J. Noninvasive biomarkers for the diagnosis and management of autoimmune hepatitis. Hepatology 2022, 76, 1862–1879.
- Kaur, N.; Goyal, G.; Garg, R.; Tapasvi, C.; Chawla, S.; Kaur, R. Potential role of noninvasive biomarkers during liver fibrosis. World J. Hepatol. 2021, 13, 1919–1935.
- Gu, J.; Liang, D.; Pierzynski, J.A.; Zheng, L.; Ye, Y.; Zhang, J.; Ajani, J.A.; Wu, X. D-mannose: A novel prognostic biomarker for patients with esophageal adenocarcinoma. Carcinogenesis 2017, 38, 162–167.
- Shah, A.K.; Hartel, G.; Brown, I.; Winterford, C.; Na, R.; Lê Cao, K.-A.; Spicer, B.A.; Dunstone, M.; Phillips, W.A.; Lord, R.V. Serum glycoprotein biomarker validation for esophageal adenocarcinoma and application to Barrett’s surveillance. bioRxiv 2018, 281220.
- Maity, A.K.; Stone, T.C.; Ward, V.; Webster, A.P.; Yang, Z.; Hogan, A.; McBain, H.; Duku, M.; Ho, K.M.A.; Wolfson, P.; et al. Novel epigenetic network biomarkers for early detection of esophageal cancer. Clin. Epigenetics 2022, 14, 23.
- Liu, Y.-Q.; Chu, L.-Y.; Yang, T.; Zhang, B.; Zheng, Z.-T.; Xie, J.-J.; Xu, Y.-W.; Fang, W.-K. Serum DSG2 as a potential biomarker for diagnosis of esophageal squamous cell carcinoma and esophagogastric junction adenocarcinoma. Biosci. Rep. 2022, 42, BSR20212612.
- Liu, W.; Wang, Q.; Chang, J.; Bhetuwal, A.; Bhattarai, N.; Zhang, F.; Tang, J. Serum proteomics unveil characteristic protein diagnostic biomarkers and signaling pathways in patients with esophageal squamous cell carcinoma. Clin. Proteom. 2022, 19, 18.
- Wei, J.; Li, R.; Lu, Y.; Meng, F.; Xian, B.; Lai, X.; Lin, X.; Deng, Y.; Yang, D.; Zhang, H.; et al. Salivary microbiota may predict the presence of esophageal squamous cell carcinoma. Genes Dis. 2022, 9, 1143–1151.
- Zhang, X.; Sun, L. Anaphylatoxin C3a: A potential biomarker for esophageal cancer diagnosis. Mol. Clin. Oncol. 2018, 8, 315–319.
- Niknejad, F.; Escriva, L.; Adel Rad, K.B.; Khoshnia, M.; Barba, F.J.; Berrada, H. Biomonitoring of Multiple Mycotoxins in Urine by GC-MS/MS: A Pilot Study on Patients with Esophageal Cancer in Golestan Province, Northeastern Iran. Toxins 2021, 13, 243.
- Okuda, Y.; Shimura, T.; Iwasaki, H.; Fukusada, S.; Nishigaki, R.; Kitagawa, M.; Katano, T.; Okamoto, Y.; Yamada, T.; Horike, S.I.; et al. Urinary microRNA biomarkers for detecting the presence of esophageal cancer. Sci. Rep. 2021, 11, 8508.
- Singh, D.; Rai, V.; Agrawal, D.K. Non-Coding RNAs in Regulating Plaque Progression and Remodeling of Extracellular Matrix in Atherosclerosis. Int. J. Mol. Sci. 2022, 23, 13731.
- Fassan, M.; Realdon, S.; Cascione, L.; Hahne, J.C.; Munari, G.; Guzzardo, V.; Arcidiacono, D.; Lampis, A.; Brignola, S.; Dal Santo, L.; et al. Circulating microRNA expression profiling revealed miR-92a-3p as a novel biomarker of Barrett’s carcinogenesis. Pathol. Res. Pract. 2020, 216, 152907.
- Chiam, K.; Wang, T.; Watson, D.I.; Mayne, G.C.; Irvine, T.S.; Bright, T.; Smith, L.; White, I.A.; Bowen, J.M.; Keefe, D.; et al. Circulating Serum Exosomal miRNAs As Potential Biomarkers for Esophageal Adenocarcinoma. J. Gastrointest. Surg. 2015, 19, 1208–1215.
- Chiam, K.; Mayne, G.C.; Wang, T.; Watson, D.I.; Irvine, T.S.; Bright, T.; Smith, L.T.; Ball, I.A.; Bowen, J.M.; Keefe, D.M.; et al. Serum outperforms plasma in small extracellular vesicle microRNA biomarker studies of adenocarcinoma of the esophagus. World J. Gastroenterol. 2020, 26, 2570–2583.
- Mayne, G.C.; Watson, D.I.; Chiam, K.; Hussey, D.J. ASO Author Reflections: Predicting the Response of Esophageal Adenocarcinoma to Chemoradiotherapy Before Surgery Using MicroRNA Biomarkers Offers Hope to Improve Outcomes by Tailoring Treatment to Predicted Responses. Ann. Surg. Oncol. 2018, 25, 755–756.
- Egyud, M.; Tejani, M.; Pennathur, A.; Luketich, J.; Sridhar, P.; Yamada, E.; Stahlberg, A.; Filges, S.; Krzyzanowski, P.; Jackson, J.; et al. Detection of Circulating Tumor DNA in Plasma: A Potential Biomarker for Esophageal Adenocarcinoma. Ann. Thorac. Surg. 2019, 108, 343–349.
- Bonazzi, V.F.; Aoude, L.G.; Brosda, S.; Lonie, J.M.; Patel, K.; Bradford, J.J.; Koufariotis, L.T.; Wood, S.; Smithers, B.M.; Waddell, N.; et al. ctDNA as a biomarker of progression in oesophageal adenocarcinoma. ESMO Open 2022, 7, 100452.
- Openshaw, M.R.; Mohamed, A.A.; Ottolini, B.; Fernandez-Garcia, D.; Richards, C.J.; Page, K.; Guttery, D.S.; Thomas, A.L.; Shaw, J.A. Longitudinal monitoring of circulating tumour DNA improves prognostication and relapse detection in gastroesophageal adenocarcinoma. Br. J. Cancer 2020, 123, 1271–1279.
- Chidambaram, S.; Markar, S.R. Clinical utility and applicability of circulating tumor DNA testing in esophageal cancer: A systematic review and meta-analysis. Dis. Esophagus 2022, 35, doab046.
- Dakubo, G.D. Esophageal Cancer Biomarkers in Circulation. In Cancer Biomarkers in Body Fluids; Springer: Cham, Switzerland, 2017; pp. 147–178.
- Ju, C.; He, J.; Wang, C.; Sheng, J.; Jia, J.; Du, D.; Li, H.; Zhou, M.; He, F. Current advances and future perspectives on the functional roles and clinical implications of circular RNAs in esophageal squamous cell carcinoma: More influential than expected. Biomark. Res. 2022, 10, 41.
- Zhang, Y.; Xu, Y.; Li, Z.; Zhu, Y.; Wen, S.; Wang, M.; Lv, H.; Zhang, F.; Tian, Z. Identification of the key transcription factors in esophageal squamous cell carcinoma. J. Thorac. Dis. 2018, 10, 148–161.
- Yao, C.; Liu, H.N.; Wu, H.; Chen, Y.J.; Li, Y.; Fang, Y.; Shen, X.Z.; Liu, T.T. Diagnostic and Prognostic Value of Circulating MicroRNAs for Esophageal Squamous Cell Carcinoma: A Systematic Review and Meta-analysis. J. Cancer 2018, 9, 2876–2884.
- Hoshino, I.; Ishige, F.; Iwatate, Y.; Gunji, H.; Kuwayama, N.; Nabeya, Y.; Yokota, H.; Takeshita, N.; Iida, K.; Nagase, H.; et al. Cell-free microRNA-1246 in different body fluids as a diagnostic biomarker for esophageal squamous cell carcinoma. PLoS ONE 2021, 16, e0248016.
- Kahng, D.H.; Kim, G.H.; Park, S.J.; Kim, S.; Lee, M.W.; Lee, B.E.; Hoseok, I. MicroRNA Expression in Plasma of Esophageal Squamous Cell Carcinoma Patients. J. Korean Med. Sci. 2022, 37, e197.
- Du, J.; Zhang, L. Analysis of salivary microRNA expression profiles and identification of novel biomarkers in esophageal cancer. Oncol. Lett. 2017, 14, 1387–1394.
- Xie, Z.J.; Chen, G.; Zhang, X.C.; Li, D.F.; Huang, J.; Li, Z.J. Saliva supernatant miR-21: A novel potential biomarker for esophageal cancer detection. Asian Pac. J. Cancer Prev. 2012, 13, 6145–6149.
- Xie, Z.; Chen, G.; Zhang, X.; Li, D.; Huang, J.; Yang, C.; Zhang, P.; Qin, Y.; Duan, Y.; Gong, B.; et al. Salivary microRNAs as promising biomarkers for detection of esophageal cancer. PLoS ONE 2013, 8, e57502.
- Li, K.; Lin, Y.; Luo, Y.; Xiong, X.; Wang, L.; Durante, K.; Li, J.; Zhou, F.; Guo, Y.; Chen, S.; et al. A signature of saliva-derived exosomal small RNAs as predicting biomarker for esophageal carcinoma: A multicenter prospective study. Mol. Cancer 2022, 21, 21.
- Parameshwaran, K.; Sharma, P.; Rajendra, S.; Stelzer-Braid, S.; Xuan, W.; Rawlinson, W.D. Circulating human papillomavirus DNA detection in Barrett’s dysplasia and esophageal adenocarcinoma. Dis. Esophagus 2019, 32, doz064.
- Kato, S.; Okamura, R.; Baumgartner, J.M.; Patel, H.; Leichman, L.; Kelly, K.; Sicklick, J.K.; Fanta, P.T.; Lippman, S.M.; Kurzrock, R. Analysis of Circulating Tumor DNA and Clinical Correlates in Patients with Esophageal, Gastroesophageal Junction, and Gastric Adenocarcinoma. Clin. Cancer Res. 2018, 24, 6248–6256.
This entry is offline, you can click here to edit this entry!
