Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by José Pérez de la Lastra.
Fish, like all other animals, are exposed to constant contact with microbes, both on their skin and on the surfaces of their respiratory and digestive systems. Fish have a system of non-specific immune responses that provides them with initial protection against infection and allows them to survive under normal conditions despite the presence of these potential invaders.
- antimicrobial peptide
- fish
- piscidin
- Teleost
- innate immunity
1. Introduction
There is growing evidence that the equilibrium of all living things, including those that live in water, depends on a constant dialog with the microbes that cover their surfaces [1]. The diversity of microorganisms in the oceans is probably underestimated. If a subset of these bacteria is indeed harmful to marine fishes, it is plausible that coevolutionary processes drove the evolution of innate antimicrobial defenses in these organisms. Consequently, it is reasonable to expect a corresponding set of such defenses in fish [2]. Fish express all major AMP families, including defensins, cathelicidins, hepcidins, and histone peptides [3]. Bony fishes are divided into two subclasses: Holostei and Teleostei, with the latter being the most important and abundant, comprising 96% of all fish species [4]. Holostei, such as North American bowfins (Amiiformes) and gars (Semionotiformes), are the closest living relatives of Teleostei. Teleosts inhabit various niches and are at increased risk of being attacked by a variety of potential pathogens [5]. The main structural difference between Teleostei and other bony fishes is in the jaw bones. Teleostei have a movable premaxilla and accompanying changes in jaw musculature that allow them to extend their jaws out of their mouths. This gives them a significant advantage, allowing them to grab their prey and guide it into their mouth. They also have similar-sized caudal fin lobes and a spine that ends at the caudal peduncle [6,7][6][7]. Under normal circumstances, fish can cope with these potential invaders thanks to a system of non-specific immune responses that provides an initial defense against pathogens. Several pathogens enter fish because their skin, gills, and intestines are the main body surfaces that come into direct contact with the environment [8]. However, the exact methods by which hosts interact with their microbiome and the ways in which the emergence of a species’ microbiome is regulated are still largely unclear [9].
The Teleost skin consists of two main parts: the epidermis and the dermis. The cells on the surface are not keratinized [10]. When the differentiation process of the mucous cells begins in the stratum germinativum, the nascent cells migrate to the skin surface and secrete their contents [11]. Fish do not have lymph nodes or other lymphatic tissue as found in mammals [12]. Most pathogens enter Teleosts through the mucous membranes, which are the main point of contact between fish and their immediate environment [13]. The cells that produce the alarm substance and melanophores are located in the dermis or deeper epidermis and do not reach the skin surface. Injury to the epidermis is the only trigger for the release of the alarm substance stored in the skin cells. Both the epidermis and dermis are necessary for skin health [14].
Although the skin, gills, and intestinal mucosa are constantly exposed to microorganisms from the environment, infections or life-threatening lesions do not occur under normal conditions [15]. Antimicrobial peptides (AMPs) are one type of innate immune protection [16,17][16][17]. In vertebrates, low-molecular-weight antibacterial peptides are typically found in peripheral blood leukocytes and on mucous membranes [8]. Similarly, mucus extracts from the skin of several fish species, including rainbow trout, have been identified as having antimicrobial peptides against selected bacteria [18]. Host defense peptides have been found to have close genetic, structural, and functional relatives in non-fish species thanks to advances in genome sequencing research [19,20][19][20]. Recent research has shown that the peptide piscidin is synthesized in the epithelia of the gills, skin, stomach, and gut of a variety of Teleost species. Piscidins are present in eosinophil cells in epithelial tissues, suggesting that they play an important role in innate defense in these tissues [21].
Silphalin and Noga discovered the first piscidin in the hybrid striped bass (Morone saxatilis × Morone chrysops) [22]. Piscidins are a family of cationic AMPs that are produced by fish and have broad-spectrum antimicrobial activity against bacteria, fungi, and viruses [23]. The piscidin family includes the well-studied peptides Epinecidin-1, Myxinidin, Chrysophin, Diacentracin, Pleurocidins, and Moronecidins, all of which play important roles in innate immunity in fish [23,24][23][24]. They are relatively short peptides, typically consisting of 22–40 amino acids, and are rich in arginine and cysteine residues [25]. Piscidins are primarily produced by fish leukocytes, including mast cells, neutrophils, and macrophages, and are stored in granules within these cells [26]. They are released in response to microbial infections or other inflammatory stimuli and act by disrupting the membranes [27]. Using immunohistochemistry, researchers identified piscidins in the tissues of fishes of the families Moronidae, Sciaenidae, Serranidae, Cichlidae, Siganidae, and Belontidae [21].
Because of their broad-spectrum antimicrobial activity and immunomodulatory effects, piscidins could be useful in treating a variety of infectious and inflammatory diseases. Researchers are also investigating the potential use of piscidins as a therapeutic agent in humans [28]. One of the most remarkable features of piscidins as potential therapeutic agents is their broad-spectrum antimicrobial activity. Studies have demonstrated the efficacy of piscidins against a wide range of microorganisms, including bacteria, fungi, and viruses, which makes them potentially useful for treating a variety of infectious diseases [29]. Another notable feature of piscidins is their ability to modulate the immune response. Piscidins have been shown to stimulate the production of cytokines and chemokines, which can help to recruit immune cells to sites of infection or injury. They can also promote wound healing and have anti-inflammatory effects [30,31][30][31]. Piscidins are relatively small peptides, which makes them easier to synthesize than larger proteins. This could make them a more cost-effective and accessible therapeutic option than other antimicrobial agents [32,33][32][33]. Resistance to antibiotics caused by their overuse has been a concern for some time. Many bacterial infections are becoming more common and widespread, with devastating effects on human health [34,35][34][35]. With their potent antimicrobial activity and distinct antimicrobial processes, piscidins could be a good substitute for current antimicrobials and offer an advantage over conventional antibiotics in the fight against drug-resistant bacterial diseases [36]. Because piscidins are naturally produced by fish, they may be less likely to cause adverse effects in humans than synthetic antimicrobial agents [37].
2. UniProt-Reviewed Piscidins
Piscidins are also called pleurocidins, in reference to one of the first AMP sequences isolated from the mucosal cells of flounder [41][38]. To date, piscidins have been characterized in a variety of Teleost species, including cod (Gadus morhua), red bream (Chrysophrys major), sea bass (Dicentrarchus labrax), grouper (Epinephelus coioides), rainbow trout (Oncorhynchus mykiss), and striped bass (Morone), to cite a few. UniProt is a database that contains extensive descriptions of proteins and their role in various biological processes, molecular interactions, and pathways, as well as links to other useful databases [43][39]. According to UniProt, the pleurocidin protein family has about 360 entries (accessed on 14 March 2023). However, only 11 of them (Table 1) were reviewed by UniProt curators (Swiss-Prot). Swiss-Prot, founded in 1986, is included in the reviewed area of the UniProt Knowledgebase. Swiss-Prot is a high-quality, manually annotated, non-redundant protein sequence database that brings together experimental results, calculated features, and scientific conclusions. The TrEMBL part of the UniProtKB database was first made available in 1996 in response to the growing influx of data that was a direct result of genomic studies. The mature peptides and pro-domains of piscidin peptides from different fish species show little similarity [44][40]. The sequences of most piscidins’ mature active peptides are often predicted via homology or alignment with already known mature peptides from other fishes [45][41]. This is an important limitation to the study of these peptide families.Table 1. Pleurocidin protein family found in InterPro database (accessed on 14 March 2023), reviewed by UniProt curators (Swiss-Prot), and length of their active peptides obtained from each entry.
| Accession | Name | Species | Length |
|---|---|---|---|
| Q90ZX8 | Pleurocidin-WF4 | Pseudopleuronectes americanus (Winter flounder) | 25 |
| P81941 | Pleurocidin | Pseudopleuronectes americanus (Winter flounder) | 25 |
| P0DUJ5 | Pteroicidin-alpha | Pterois volitans (Red lionfish) | 21 |
| P0C006 | Piscidin-3 | Morone chrysops × Morone saxatilis (White bass × Striped bass) | 22 |
| Q8UUG0 | Moronecidin Ms | Morone saxatilis (Striped bass) | 22 |
| P59906 | Dicentracin | Dicentrarchus labrax (European seabass) | 22 |
| Q8UUG2 | Moronecidin Mc | Morone chrysops (White bass) | 22 |
| Q90VW7 | Pleurocidin-WF3 | Pseudopleuronectes americanus (Winter flounder) | 25 |
| Q90VW7 | Chrysophsin-3 | Pagrus major (Red seabream) | 20 |
| P83546 | Chrysophsin-3 | Pagrus major (Red seabream) | 25 |
| P83545 | Chrysophsin-1 | Pagrus major (Red seabream) | 25 |
3. Evolutionary Diversity of Piscidins
Piscidin is a class of peptides that is one of the most abundant AMPs in fish. In the course of studying AMPs unique to fish, wresearchers now know that certain peptides, originally named pleurocidins, and which weresearchers call piscidins, are present in a number of Teleost species from several families [21]. Piscidins are an extremely diverse family of AMPs, each with its own unique amino acid sequence. It was found that the piscidin peptides studied in detail varied in length and amino acid sequence depending on the fish species studied [44][40]. Piscidins are subject to both positive Darwinian selection and gene duplication, which would explain the wide range of peptides and low degree of sequence similarity among members of the piscidin family [52][48]. This suggests that various ecological and evolutionary influences have affected the evolution of piscidin peptides in different fish species [52][48]. Although there is some homology among piscidin peptides, variations in sequence and structure suggest that different piscidin peptides have evolved to perform specialized functions in different fish species [8]. Using the Hidden Markov Model and Seeded Guide Tree methods, the multiple sequence alignment tool Clustal Omega (accessed on 14 March 2023) created alignments between three or more sequences [53][49]. Asterisks denote identical residue, a colon indicates strong homology, and a period indicates weak homology, respectively, based on the Gonnet Pam250 matrix. The alignment of the 11 piscidin peptides shows that certain residues are conserved in two locations (squares 1 and 2 of Figure 1).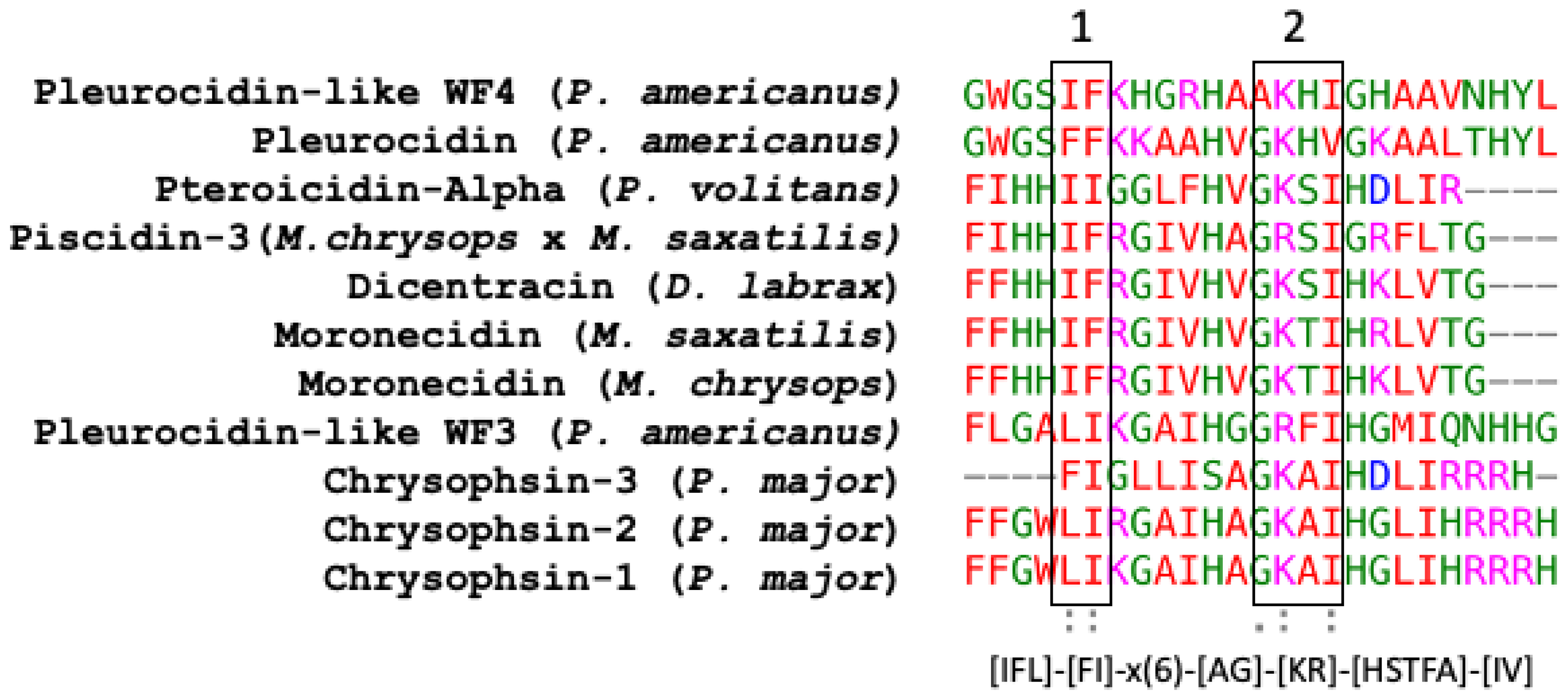
Figure 1. Alignment of the amino acid sequences of the mature active peptides from UniProt-reviewed piscidins (pleurocidin) using Clustal Omega. A colon (:) indicates strong homology, and a period (.) indicates weak homology, respectively, based on the Gonnet Pam250 matrix.
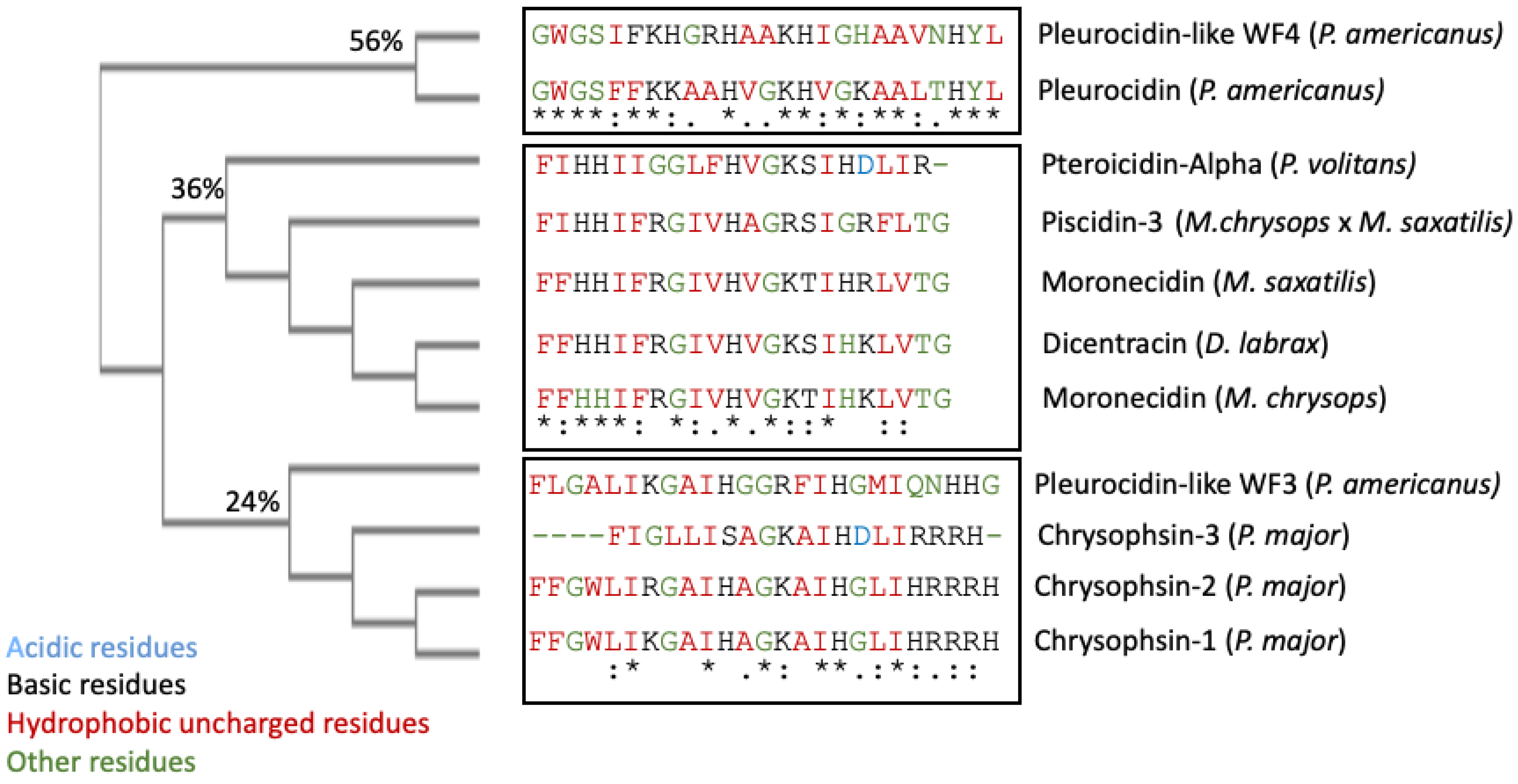
Figure 2. Clustal Omega guide tree obtained via sequence similarity of UniProt-reviewed mature piscidins. The percentages of identity among the clusters are shown in each branch. Acidic residues are colored in blue, basic residues in black, hydrophobic uncharged residues in red, and other residues in green, respectively. Asterisks (*) denote identical residues, a colon (:) indicates strong homology, and a period (.) indicates weak homology, respectively.
4. Piscidin Gene Arrangement, Processing, and Expression
Basal expression patterns of piscidin genes have been found to differ both within and between fish species. Furthermore, the expression levels of the different isoforms can vary widely within a species. Piscidin expression also begins early in fish development and continues to increase throughout the life cycle. For example, transcripts of pleurocidin-like genes have been found in winter flounder larvae as early as five days after hatching, and different pleurocidin-like genes are probably expressed at different developmental stages. Gill, skin, colon, brain, kidney, and spleen are just some of the tissues where these genes are consistently expressed [27]. In terms of cell types, piscidins are expressed by mast cells, rod cells, phagocytic granulocytes, and eosinophilic granulocytes. [54,55][50][51]. Piscidin gene expression has been shown to be altered following infection with a variety of pathogens [23]. Particularly, mucosal tissue contains piscidin peptides at levels that are lethal to pathogens [8]. Most piscidin genes encode a precursor with a signal peptide with 22 residues, a mature (active) peptide with 22–25 residues, and a variable C-terminal region [52,56][48][52]. Piscidins are encoded by four exons and three introns. The exons encode the signal peptide, full-length peptide, and propeptide. The 5′ untranslated region contributes to the formation of the first exon, which continues to the first nucleotide of the second exon. Exon 2 encodes the signal peptides, while exons 2, 3, and 4 encode the mature peptides. Exon 4 encodes the pro-domains, followed by the 3′ UTR. Exons in piscidin genes vary in size, with exon 4 being the largest [44][40]. Figure 3 shows, as an example, the gene organization of the Dicentracin peptide with four exons and three introns.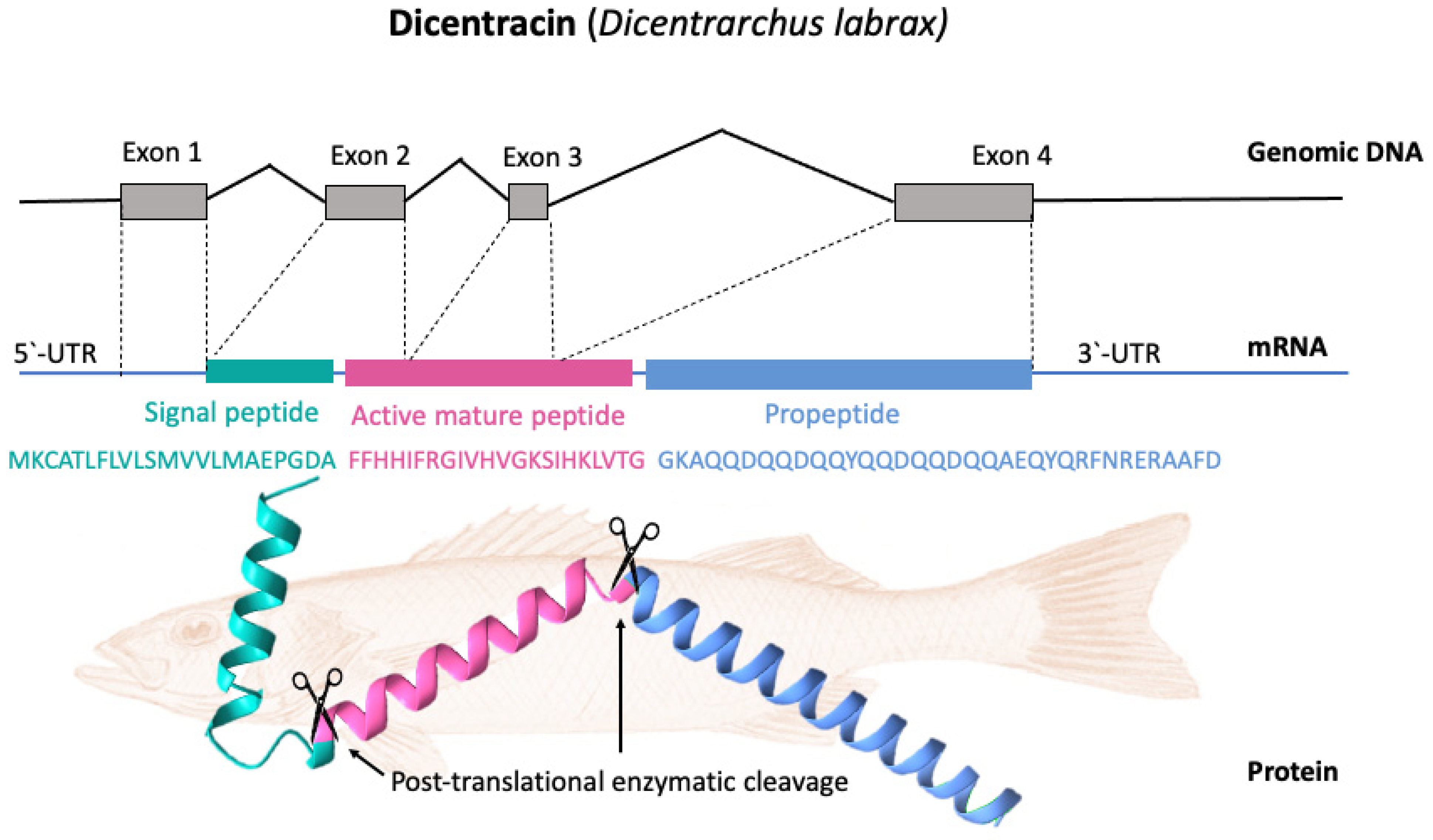
Figure 3. Gene organization and processing of Dicentracin piscidin from Dicentrarchus labrax. The precursor protein consists of a signal peptide, the active peptide, and the propeptide. The active mature Dicentracin peptide is obtained via post-translational cleavage of the pre-peptide.
References
- Chen, Y.E.; Fischbach, M.A.; Belkaid, Y. Skin microbiota–host interactions. Nature 2018, 553, 427–436.
- Patrzykat, A.; Hancock, R.E. Host defense peptides in fish: From the peculiar to the mainstream. In Fish Defenses; CRC Press: Boca Raton, FL, USA, 2019; pp. 43–61.
- Masso-Silva, J.A.; Diamond, G. Antimicrobial Peptides from Fish. Pharmaceuticals 2014, 7, 265–310.
- Ravi, V.; Venkatesh, B. The divergent genomes of teleosts. Annu. Rev. Anim. Biosci. 2018, 6, 47–68.
- Wootton, R.J. Ecology of Teleost Fishes; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; Volume 1.
- Arratia, G. Complexities of early Teleostei and the evolution of particular morphological structures through time. Copeia 2015, 103, 999–1025.
- Witten, P.E.; Hall, B.K. Teleost skeletal plasticity: Modulation, adaptation, and remodelling. Copeia 2015, 103, 727–739.
- Gomez, D.; Sunyer, J.O.; Salinas, I. The mucosal immune system of fish: The evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 2013, 35, 1729–1739.
- Kelly, C.; Salinas, I. Under pressure: Interactions between commensal microbiota and the teleost immune system. Front. Immunol. 2017, 8, 559.
- Xu, Z.; Parra, D.; Gómez, D.; Salinas, I.; Zhang, Y.-A.; von Gersdorff Jørgensen, L.; Heinecke, R.D.; Buchmann, K.; LaPatra, S.; Sunyer, J.O. Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. Proc. Natl. Acad. Sci. USA 2013, 110, 13097–13102.
- Ángeles Esteban, M. An overview of the immunological defenses in fish skin. Int. Sch. Res. Not. 2012, 2012, 853470.
- Kogame, T.; Kabashima, K.; Egawa, G. Putative immunological functions of inducible skin-associated lymphoid tissue in the context of mucosa-associated lymphoid tissue. Front. Immunol. 2021, 12, 733484.
- Dickerson, H.W. The biology of teleost mucosal immunity. Fish Def. Pathog. Parasites Predat. 2009, 2, 1–42.
- Fontenot, D.K.; Neiffer, D.L. Wound management in teleost fish: Biology of the healing process, evaluation, and treatment. Vet. Clin. Exot. Anim. Pract. 2004, 7, 57–86.
- Watts, M.; Munday, B.; Burke, C. Immune responses of teleost fish. Aust. Vet. J. 2001, 79, 570–574.
- Ellis, A. Innate host defense mechanisms of fish against viruses and bacteria. Dev. Comp. Immunol. 2001, 25, 827–839.
- Magnadóttir, B. Innate immunity of fish (overview). Fish Shellfish Immunol. 2006, 20, 137–151.
- Plouffe, D.A.; Hanington, P.C.; Walsh, J.G.; Wilson, E.C.; Belosevic, M. Comparison of select innate immune mechanisms of fish and mammals. Xenotransplantation 2005, 12, 266–277.
- Salger, S.A.; Cassady, K.R.; Reading, B.J.; Noga, E.J. A diverse family of host-defense peptides (piscidins) exhibit specialized anti-bacterial and anti-protozoal activities in fishes. PLoS ONE 2016, 11, e0159423.
- Priyam, M.; Bhat, R.A.; Kumar, N. Recent Advances in Antimicrobial Peptides to Improve Fish Health. In Biotechnological Advances in Aquaculture Health Management; Gupta, S.K., Giri, S.S., Eds.; Springer: Singapore, 2021; pp. 165–187.
- Silphaduang, U.; Colorni, A.; Noga, E. Evidence for widespread distribution of piscidin antimicrobial peptides in teleost fish. Dis. Aquat. Org. 2006, 72, 241–252.
- Noga, E.; Fan, Z.; Silphaduang, U. Histone-like proteins from fish are lethal to the parasitic dinoflagellate Amyloodinium ocellatum. Parasitology 2001, 123, 57–65.
- Raju, S.V.; Sarkar, P.; Kumar, P.; Arockiaraj, J. Piscidin, fish antimicrobial peptide: Structure, classification, properties, mechanism, gene regulation and therapeutical importance. Int. J. Pept. Res. Ther. 2021, 27, 91–107.
- Hazam, P.K.; Chen, J.-Y. Therapeutic utility of the antimicrobial peptide Tilapia Piscidin 4 (TP4). Aquac. Rep. 2020, 17, 100409.
- Valero, Y.; Saraiva-Fraga, M.; Costas, B.; Guardiola, F.A. Antimicrobial peptides from fish: Beyond the fight against pathogens. Rev. Aquac. 2020, 12, 224–253.
- Mulero, I.; Noga, E.J.; Meseguer, J.; García-Ayala, A.; Mulero, V. The antimicrobial peptides piscidins are stored in the granules of professional phagocytic granulocytes of fish and are delivered to the bacteria-containing phagosome upon phagocytosis. Dev. Comp. Immunol. 2008, 32, 1531–1538.
- Katzenback, B.A. Antimicrobial peptides as mediators of innate immunity in teleosts. Biology 2015, 4, 607–639.
- Conlon, J.M. Host-defense peptides of the skin with therapeutic potential: From hagfish to human. Peptides 2015, 67, 29–38.
- Shabir, U.; Ali, S.; Magray, A.R.; Ganai, B.A.; Firdous, P.; Hassan, T.; Nazir, R. Fish antimicrobial peptides (AMP’s) as essential and promising molecular therapeutic agents: A review. Microb. Pathog. 2018, 114, 50–56.
- Mokhtar, D.M.; Zaccone, G.; Alesci, A.; Kuciel, M.; Hussein, M.T.; Sayed, R.K. Main Components of Fish Immunity: An Overview of the Fish Immune System. Fishes 2023, 8, 93.
- Valero, Y.; Chaves-Pozo, E.; Meseguer, J.; Esteban, M.A.; Cuesta, A. Biological role of fish antimicrobial peptides. Antimicrob. Pept. 2013, 2, 31–60.
- Paria, A.; Vinay, T.; Gupta, S.K.; Choudhury, T.G.; Sarkar, B. Antimicrobial peptides: A promising future alternative to antibiotics in aquaculture. World Aquac. 2018, 49, 67–69.
- Hancock, R.E.; Alford, M.A.; Haney, E.F. Antibiofilm activity of host defence peptides: Complexity provides opportunities. Nat. Rev. Microbiol. 2021, 19, 786–797.
- Pérez de la Lastra, J.M.; Anand, U.; González-Acosta, S.; López, M.R.; Dey, A.; Bontempi, E.; Morales delaNuez, A. Antimicrobial resistance in the COVID-19 landscape: Is there an opportunity for anti-infective antibodies and antimicrobial peptides? Front. Immunol. 2022, 13, 921483.
- Bharadwaj, A.; Rastogi, A.; Pandey, S.; Gupta, S.; Sohal, J.S. Multidrug-Resistant Bacteria: Their mechanism of action and prophylaxis. BioMed Res. Int. 2022, 2022, 5419874.
- Sinha, R.; Shukla, P. Antimicrobial peptides: Recent insights on biotechnological interventions and future perspectives. Protein Pept. Lett. 2019, 26, 79–87.
- Chaturvedi, P.; Bhat, R.A.H.; Pande, A. Antimicrobial peptides of fish: Innocuous alternatives to antibiotics. Rev. Aquac. 2020, 12, 85–106.
- Douglas, S.; Gallant, J.; Gong, Z.; Hew, C. Cloning and developmental expression of a family of pleurocidin-like antimicrobial peptides from winter flounder, Pleuronectes americanus (Walbaum). Dev. Comp. Immunol. 2001, 25, 137–147.
- Consortium, T.U. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2020, 49, D480–D489.
- Barroso, C.; Carvalho, P.; Carvalho, C.; Santarém, N.; Gonçalves, J.F.M.; Rodrigues, P.N.S.; Neves, J.V. The Diverse Piscidin Repertoire of the European Sea Bass (Dicentrarchus labrax): Molecular Characterization and Antimicrobial Activities. Int. J. Mol. Sci. 2020, 21, 4613.
- Buonocore, F.; Randelli, E.; Casani, D.; Picchietti, S.; Belardinelli, M.C.; de Pascale, D.; De Santi, C.; Scapigliati, G. A piscidin-like antimicrobial peptide from the icefish Chionodraco hamatus (Perciformes: Channichthyidae): Molecular characterization, localization and bactericidal activity. Fish Shellfish Immunol. 2012, 33, 1183–1191.
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427.
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354.
- Sharma, P.K.; Yadav, I.S. Biological databases and their application. In Bioinformatics; Elsevier: Amsterdam, The Netherlands, 2022; pp. 17–31.
- Mulder, N.J.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Buillard, V.; Cerutti, L.; Copley, R. New developments in the InterPro database. Nucleic Acids Res. 2007, 35, D224–D228.
- Qiao, Y.; Ma, X.; Zhang, M.; Zhong, S. Cerocin, a novel piscidin-like antimicrobial peptide from black seabass, Centropristis striata. Fish Shellfish Immunol. 2021, 110, 86–90.
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009, 37, D211–D215.
- Fernandes, J.M.; Ruangsri, J.; Kiron, V. Atlantic cod piscidin and its diversification through positive selection. PLoS ONE 2010, 5, e9501.
- Sievers, F.; Higgins, D.G. The clustal omega multiple alignment package. Mult. Seq. Alignment Methods Protoc. 2021, 2231, 3–16.
- Ruangsri, J.; Salger, S.A.; Caipang, C.M.; Kiron, V.; Fernandes, J.M. Differential expression and biological activity of two piscidin paralogues and a novel splice variant in Atlantic cod (Gadus morhua L.). Fish Shellfish Immunol. 2012, 32, 396–406.
- Serna-Duque, J.A.; Cuesta, A.; Sánchez-Ferrer, Á.; Esteban, M.Á. Two duplicated piscidin genes from gilthead seabream (Sparus aurata) with different roles in vitro and in vivo. Fish Shellfish Immunol. 2022, 127, 730–739.
- Zaccone, G.; Capillo, G.; Fernandes, J.M.O.; Kiron, V.; Lauriano, E.R.; Alesci, A.; Lo Cascio, P.; Guerrera, M.C.; Kuciel, M.; Zuwala, K. Expression of the antimicrobial peptide piscidin 1 and neuropeptides in fish gill and skin: A potential participation in neuro-immune interaction. Mar. Drugs 2022, 20, 145.
- Wiederanders, B.; Kaulmann, G.; Schilling, K. Functions of propeptide parts in cysteine proteases. Curr. Protein Pept. Sci. 2003, 4, 309–326.
- Xhindoli, D.; Pacor, S.; Benincasa, M.; Scocchi, M.; Gennaro, R.; Tossi, A. The human cathelicidin LL-37—A pore-forming antibacterial peptide and host-cell modulator. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2016, 1858, 546–566.
- Umasuthan, N.; Mothishri, M.; Thulasitha, W.S.; Nam, B.-H.; Lee, J. Molecular, genomic, and expressional delineation of a piscidin from rock bream (Oplegnathus fasciatus) with evidence for the potent antimicrobial activities of Of-Pis1 peptide. Fish Shellfish Immunol. 2016, 48, 154–168.
- Go, H.-J.; Kim, C.-H.; Park, J.B.; Kim, T.Y.; Lee, T.K.; Oh, H.Y.; Park, N.G. Biochemical and molecular identification of a novel hepcidin type 2-like antimicrobial peptide in the skin mucus of the pufferfish Takifugu pardalis. Fish Shellfish Immunol. 2019, 93, 683–693.
More
