Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Dominika Petríková and Version 2 by Rita Xu.
Deep learning (DL) and convolutional neural networks (CNNs) have achieved state-of-the-art performance in many medical image analysis tasks. Histopathological images contain valuable information that can be used to diagnose diseases and create treatment plans. Therefore, the application of DL for the classification of histological images is a rapidly expanding field of research.
- classification
- convolutional neural networks
- deep learning
1. Introduction
Traditionally, pathology diagnosis has been performed by a human pathologist observing stained specimens from tumors on glass slides using a microscope to diagnose cancer. In recent years, deep learning has rapidly developed, and more and more entire tissue slides are being captured digitally by scanners and saved as whole slide images (WSIs) [1]. Since a large amount of WSIs are being digitized, it is only natural that many attempts have been made to explore the potential of deep learning on histopathological image analysis. Histological images and tasks have unique characteristics, and specific processing techniques are often required [2].
2. Convolutional Neural Network
Neural networks are the foundation of most DNN algorithms, consisting of interconnected units called neurons organized into layers, including input, hidden, and output layers. DNNs have multiple hidden layers. A neuron’s output, or activation, is a linear combination of its inputs and parameters (weights and bias) transformed by an activation function. Common activation functions in neural networks include sigmoid, hyperbolic tangent, and ReLU functions. At the final output layer, activations are mapped to a distribution over classes using the softmax function [3][4][6,7]. One of the most popular and commonly used supervised deep learning networks is CNNs, which are often employed for visual data processing of images and video sequences [5][6][7][8,9,10]. CNNs consist of three types of layers: convolutional layers, pooling layers, and fully connected layers, as shown in Figure 1. The convolutional layer is the most significant component of the CNN architecture. It consists of several filters, also called kernels, which are represented as a grid of discrete values. These values are referred to as kernel weights and are tuned during the training phase. The convolution operation consists of the kernel sliding over the whole image horizontally and vertically. Additionally, the dot product is calculated between the image and kernel by multiplying corresponding values and summing up to create a scalar value at each position. In particular, each kernel is convolved over the input matrix to obtain a feature map. Subsequently, the feature maps generated by the convolutional operation are sub-sampled in the pooling layer. The convolution and pooling layers together form a pipeline called feature extraction. Above all, the fully connected layers combine the features extracted by the previous layers to perform the final classification task [5][8][9][8,11,12].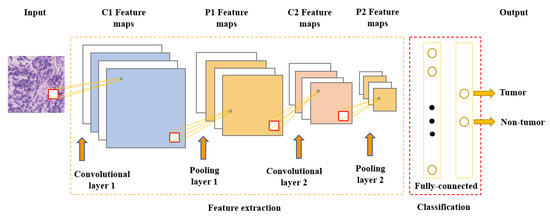
Figure 1. Convolutional neural network architecture.
3. Classification of Histopathology Images
This section provides a general overview of recent publications using deep learning and convolutional neural networks (CNNs) in digital pathology. The focus of this work is solely on supervised learning tasks applied for the classification of histological images. This category includes models that perform image-level classification, such as tumor subtype classification and grading, or use a sliding window approach to identify tissue types. Most deep learning approaches do not use the whole-slide image (WSI) as input because it would be computationally expensive (high dimensionality). Instead, they extract small square patches and assign a label to them. Existing methods can be grouped according to the level of annotations they employ. Based on the type of annotations used for training, two subcategories may be identified: the strong-annotations approach (patch-level annotations) and the weak-annotations approach (slide-level annotations) [10][13]. The first approach relies on the identification of regions of interest and the detailed localization of tumors by certified pathologists, while for the latter approach, it is sufficient to assign a specific class to a whole-slide image. In this work, a survey of the strong-annotations approach is conducted.
3.1. Strong-Annotations Approach (Patch-Level Annotation)
Referring to patch-level annotations as strong means that all extracted patches have their own label class. Typically, patch labels are derived from pixel-level annotations. Manually annotating pixels is very time-consuming and laborious work requiring an expert approach. For instance, pathologists have to localize and annotate all pixels or cells in WSI by contouring the whole tumor. This approach is shown in Figure 2. Therefore, there are currently very few strongly annotated histological images. Besides whole-slide image classification, pixel-wise/patch-wise predictions with the sliding window method enable spatial predictions such as localization and detection of cancerous cells/tissue. In addition, stacking patch predictions next to each other builds a WSI heatmap, so the model can be considered interpretable. Multiple examples of using CNNs in the problem of patch classification employ a single-stage approach when the patch is classified using one CNN architecture. In contrast, several approaches use a multi-stage workflow, where typically the output of one CNN architecture is fed into another CNN that delivers the final decision. Of course, even more CNN models can be included in such a workflow that can be labeled as multi-stage classification. For the one-stage approach, one can differentiate between models that have been trained from scratch with artificially initiated weights and models that use pre-trained CNN architectures on data often not related to the original problem. For multi-stage problems, such differentiation becomes difficult due to many possibilities, since some CNNs from the multi-stage workflow may be trained from scratch, while others may be pre-trained. In Figure 3, the top graphic shows the categorization of CNN methods used in this section.
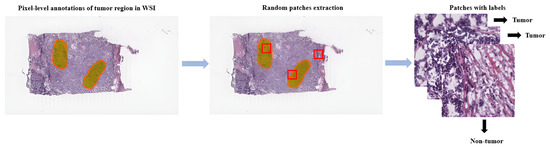
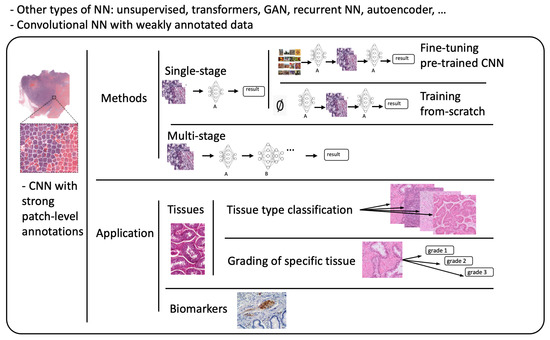

Figure 2. Construction of patches from pixel-level annotations of WSI.

Figure 3. Methods: Categorization of CNN methods. Application: Categorization of application areas.
3.2. Fine-Tuning
The easiest way of training CNNs with a limited amount of data is using one of the well-known pre-trained architectures. Typically, models are initialized using weights pre-trained on ImageNet and fine-tuned on histopathological images. In [11][14], the authors fine-tuned VGGNet [12][15], ResNet [13][16], and InceptionV4 [14][17] models to obtain the probabilities of small patches (100 × 100 pixels), being tumor-infiltrating lymphocyte (TIL)-positive or TIL-negative extracted from WSIs of 23 cancer types. For the region classification performance, they extracted bigger super-patches (800 × 800 pixels) and annotated them with three categories (Low TIL, Medium TIL, or High TIL) based on the ratio of TIL-positive area. To obtain a prediction of the category, super-patches were divided into an 8x8 grid and each square (100 × 100 pixel patch) was classified as TIL-positive or TIL-negative. Subsequently, the correlation between the score of CNN (number of positive patches in super-patch) and pathologists’ annotations was observed. In [15][18], they developed a deep learning-based six-type classifier for the identification of a wider spectrum of lung lesions including lung cancer. Furthermore, they also included pulmonary tuberculosis and organizing pneumonia, which often needs to be surgically inspected to be differentiated from cancer. EfficientNet [16][19] and ResNet were employed to carry out patch-level classification. To aggregate patch predictions into slide-level classification, two methods were compared: majority voting and mean pooling. Moreover, two-stage aggregation was implemented to prioritize cancer tissues in slides.
In [17][20], scholars proposed three steps to develop an AI-based screening method for lymph node metastases. First, they trained a segmentation model to obtain lymph node tissue from WSI and broke it into patches. Next, they used a fine-tuned Xception model to classify patches into metastasis-positive/negative. Finally, the absence or presence of two connected patches classified as positive determined the final result of WSI. In [18][21], the authors compared the accuracies of stand-alone VGG-16 and VGG-19 models with ensemble models consisting of both architectures in classifying breast cancer histopathological images as carcinoma and non-carcinoma. In [19][22], the authors compared the performance of the VGG19 architecture with methods used in supervised learning with weakly labeled data to classify ovarian carcinoma histotype. The problem of binary classification into benign and malignant lesions, with subsequent division into eight subtypes with modified EfficientNetV2 architecture on images from the BreakHis dataset, was addressed by the authors in [20][23]. Similarly, Xception was employed in [21][24] for subtyping breast cancer into four categories. The binary subtype classification of eyelid carcinoma was performed in [22][25]. They used DenseNet-161 to make predictions for every patch in WSI and then used a patch voting strategy to decide the WSI subtype. In [23][26], the authors used AlexNet [24][27], GoogLeNet [25][28], and VGG-16 to detect histopathology images with cancer cells and to classify ovarian cancer grade. Since neural networks behave like black-box models, the authors employed the Grad-CAM method to demonstrate that CNN models attended to the cancer cell organization patterns when differentiating histopathology tumor images of different grades. Grad-CAM was also employed in [26][29], where the authors used this method to provide interpretability and approximate visual diagnosis for the presentation of the model’s results to pathologists. The model consisted of three neural networks fine-tuned on a custom dataset to classify H&E stained tissue patches into five types of liver lesions, cirrhosis, and nearly normal tissue. A decision algorithm consisting of three networks was also proposed in [27][30] to detect odontogenic cyst recurrence using binary classifiers. The procedure consisted of letting the first two models make predictions. If the predictions did not match, a third model was loaded to obtain the final decision. Another example of using Grad-CAM is [28][31] to visualize classification results of the VGG16 network in grading bladder non-invasive carcinoma.
Hematoxylin-eosin (H&E) is considered as the gold standard for evaluating many cancer types. However, it contains only basic morphological information. In clinical practice, to obtain molecular information, immunohistochemical (IHC) staining is often employed. Such staining can visualize the expressions of different proteins (e.g., Ki67) on the cell membrane or nucleus. This approach is referred to as double staining. Many recent studies have shown that there is a correlation between H&E and IHC staining [29][30][31][32,33,34].
In [32][35], the authors addressed the problem of double staining in determining the number of Ki67-positive cells for cancer treatment. They employed matching pairs of IHC- and H&E-stained images and fine-tuned ResNet-18 at the cell-level from H&E images. Subsequently, to create a heat map, they transformed the CNN into a fully convolutional network without fully connected layers. As a result, the fine-tuned ResNet-18 was able to handle WSI as input and produce a heat map as output.
In [33][36], the authors proposed a modified Xception network called HE-HER2Net by adding global average pooling, batch normalization layers, dropout layers, and dense layers with a Swish activation function. The network was designed to classify H&E images into four categories based on Human epidermal growth factor receptor 2 (HER2) positivity from 0 to 3+. In addition to routine model evaluation, the authors compared their modified network to other existing architectures and claimed that HE-HER2Net surpassed all existing models in terms of accuracy, precision, recall, and AUC score.
To produce accurate models capable of generalization, it is essential to obtain large amounts of diversified data. Typically, this problem is addressed by pooling all necessary data to a centralized location. However, due to the nature of medical data, this approach has many obstacles regarding privacy and data ownership, as well as various regulatory policies (e.g., the General Data Protection Regulation GDPR of the European Union [34][37]). The authors of [35][38] simulated a Federated Learning (FL) environment to train a deep learning model that classifies cells and nuclei to identify TILs in WSI. They generated a dataset from WSIs of cancer from 12 anatomical sites and partitioned it into eight different nodes. To evaluate the performance of FL, they also trained a CNN using a centralized approach and compared the results. The study shows that the FL approach achieves similar performance to the model trained with data pooled at a centralized location.
3.3. Training from Scratch
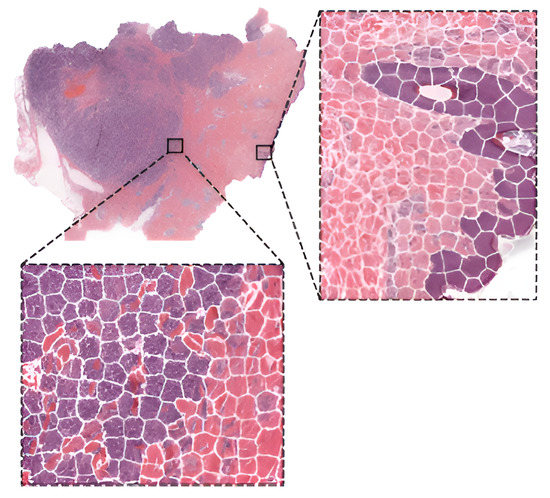
As already stated, fine-tuning is a promising method for training deep neural networks. On the other hand, it can only be applied to well-known architectures that are already pre-trained. When designing a custom CNN architecture, it needs to be trained from scratch. In [36][39], the authors proposed a method based on CNN with residual blocks (Res-Net) referred to as DeepLRHE to predict lung cancer recurrence and the risk of metastasis. Later in [37][40], scholars established the new DeepIMHL model consisting of CNN and Res-Net to predict mutated genes as biomarkers for targeted-drug therapy of lung cancer. In addition, the authors in [38][41] trained and optimized EfficientNet models on images of non-Hodgkin lymphoma and evaluated its potential to classify tumor-free reference lymph nodes, nodal small lymphocytic lymphoma/chronic lymphocytic leukemia, and nodal diffuse large B-cell lymphoma. In [39][42], the authors proposed three architectures of ResNet differing in the construction of residual blocks trained from scratch. Their suggested model achieved accuracy comparable to other state-of-the-art approaches in the classification of oral cancer histological images into three stages. To classify kidney cancer subtypes, in [40][43] the authors developed an ensemble-pyramidal model consisting of three CNNs that process images of different sizes. The authors in [41][44] demonstrated that CNN-based DL can predict the gBRCA mutation status from H&E-stained WSIs in breast cancer. According to researchers in [42][45], CNN can be employed to differentiate non-squamous Non-Small Cell Lung Cancer versus squamous cell carcinoma. To classify the tumor slide, they pooled information using the max-pooling strategy. Moreover, they added quality check with a threshold for predictions to select only tiles with a high prediction level. Additionally, to improve the prediction, they also used a virtual tissue microarray (circle from the centroid based on the pathologist’s hand-drawn tumor annotations) instead of WSI. To compare the performance of pre-trained networks with the custom ones trained from scratch, researchers in [43][46] used images of three cancer types: melanoma, breast cancer, and neuroblastoma. Unlike others using patches, the authors applied the simple linear iterative clustering (SLIC) to segment images into superpixels which group together similar neighboring pixels, as shown in Figure 4. Thus, these superpixels were classified into multiple subtype categories based on the type of cancer. To make WSI-level predictions, they used multiple specific quantification metrics such as stroma-to-tumor ratio. Although the custom NN achieved comparable results, pre-trained networks performed better on all three cancer types. A similar comparison was carried out in [44][47] for the classification of subtypes in lung cancer biopsy slides. Results showed that a CNN model built from scratch fitted to the specific pathological task could produce better performances than fine-tuning pre-trained CNNs. A comparison of training from scratch versus transfer learning was performed in [45][48]. The authors compared three approaches for training the VGG16 network: training from scratch, transfer learning as a feature extractor, and fine-tuning on images of breast cancer to detect Invasive Ductal Carcinoma. According to the results, the model trained from scratch achieved better results in terms of accuracy (0.85). However, using transfer learning, they were able to train a comparable model (accuracy 0.81) ten times faster. Furthermore, among the transfer learning approaches, transfer learning via feature extraction (accuracy 0.81), which involved retraining some of the convolutional blocks, yielded better results in less time compared to transfer learning via fine-tuning (accuracy 0.51).
Figure 4. WSI image segmentation using the SLIC superpixels algorithm.
