DNA nanotechnology has significantly advanced and might be used in biomedical applications, drug delivery, and cancer treatment. DNA nanomaterials are widely used in biomedical research involving biosensing, bioimaging, and drug delivery since they are remarkably addressable and biocompatible. Gradually, modified nucleic acids have begun to be employed to construct multifunctional DNA nanostructures with a variety of architectural designs. Aptamers are single-stranded nucleic acids (both DNAs and RNAs) capable of self-pairing to acquire secondary structure and of specifically binding with the target. Diagnosis and tumor therapy are prospective fields in which aptamers can be applied. Many DNA nanomaterials with three-dimensional structures have been studied as drug delivery systems for different anticancer medications or gene therapy agents. Different chemical alterations can be employed to construct a wide range of modified DNA nanostructures. Chemically altered DNA-based nanomaterials are useful for drug delivery because of their improved stability and inclusion of functional groups.
- DNA-based nanomaterials
- tetrahedron
- origami
- nanotube
- aptamer
- drug delivery
- endocytosis
1. Introduction
2. DNA-Based Materials
2.1. DNA Tetrahedron
Since the one-step synthesis of a tetrahedron (TDN) was first introduced, it has become one of the most widely used DNA nanostructures in biomedicine. TDNs, like other DNA nanostructures, are formed by mixing all four DNA oligonucleotides during a thermal annealing process [14][9]. Since TDN can be modified with various agents and has good biocompatibility, the use of TDN can increase the effectiveness of off-target anticancer drugs [15][10]. According to Fan et al., TDNs may successfully enter the dermis layer of mice and people’s skin and load and distribute the drug doxorubicin (DOX) to subcutaneous tumor locations [16][11]. The majority of TDNs used today are duplexes and double bundles, duplexes receiving the most research attention. TDNs can be modified with fluorescent dyes, bioligans, proteins, chemotherapy drugs, and various types of nucleic acids. Based on the various positions of the added functional groups or molecules, there are four major ways to modify TDN: vertex modification, mosaic modification, capsule modification, and cantilever modification (Figure 1) [17][12].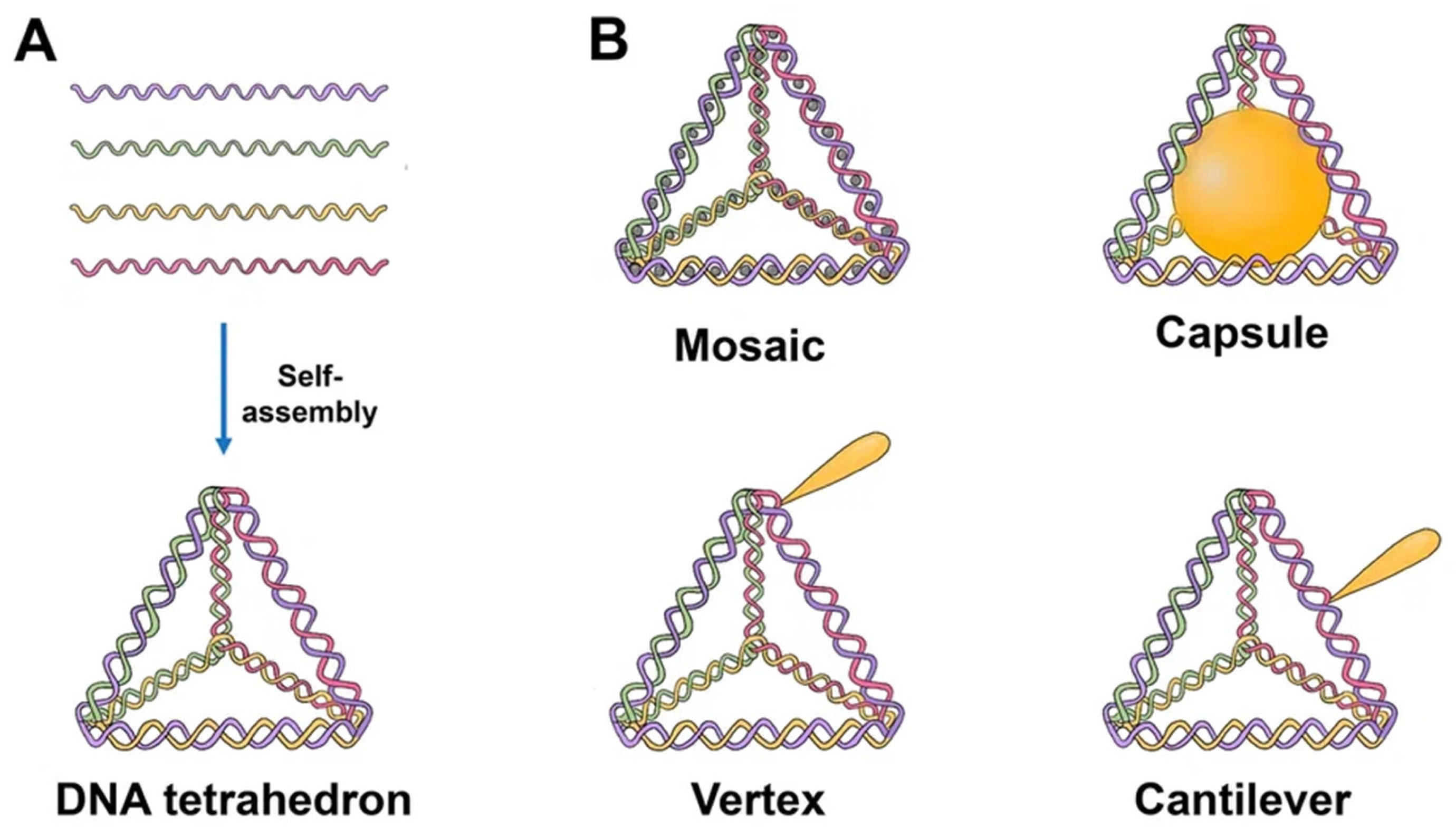
2.2. DNA Origami
Paul Rotumend introduced the concept of DNA origami technology in 2006. DNA origami is a self-assembling nanostructure in which hundreds of short “staple” oligonucleotides fold a long single-stranded DNA “scaffold” into multilayer DNA assemblies [28][23]. The scaffold is folded into a predefined form using approximately 200 complimentary short strands (“staples”) by forming crossovers at every DNA helical turn. During thermal annealing, each staple specifically links various scaffold components together according to its own sequence, folding the scaffold into the desired form (Figure 2). The advantage of using DNA origami is concluded in the addressability of specific structural components with subnanometer accuracy and precision by changing individual staples [29][24].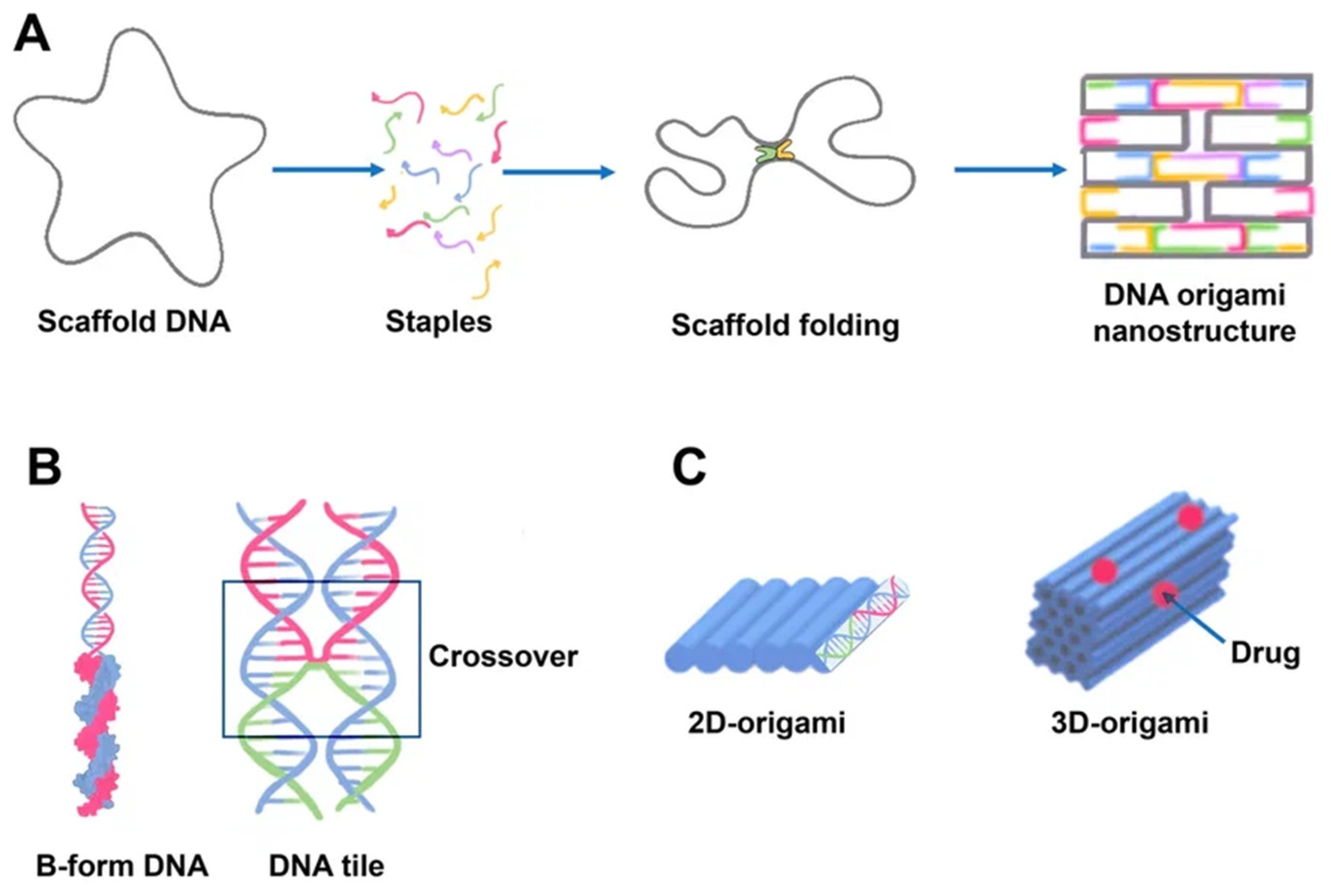
2.3. DNA Nanotube
DNA nanotubes are crystalline self-assemblies made of 10 nm-diameter DNA tiles that can grow to tens of micrometers in length. The rigidity of DNA nanotubes is hundreds of times greater than that of double-stranded DNA in general [40][35]. DNA nanotubes are special 3D structures that have great promise for biomedical applications, such as filament supporting tracks and cargo transporting carriers [41][36]. Regardless of how they are assembled, DNA nanotubes can have well-defined or indefinite lengths. Single-stranded DNA tiles, multi-crossover tiles, DNA origami, and multi-rung design methodologies for DNA nanotubes are the key assembly techniques. Tile-based motifs have been employed as DNA nanostructure building blocks (Figure 4A) since the 1990s. Recently, single-stranded tile-based 2D and 3D DNA bricks were also developed. Self-assembly of DNA nanotubes is often achieved by heating and cooling mixtures of DNA strands with a given sequence, even though the complexity of DNA nanotubes varies. Importantly, the invention of 3D DNA nanostructure assembly helped the development and use of DNA nanotubes [42][37]. Typically, DNA nanotubes are created by vertically aligning DNA duplexes into a curved motif, then closing it, or by rolling and cyclizing a two-dimensional DNA origami array [43][38].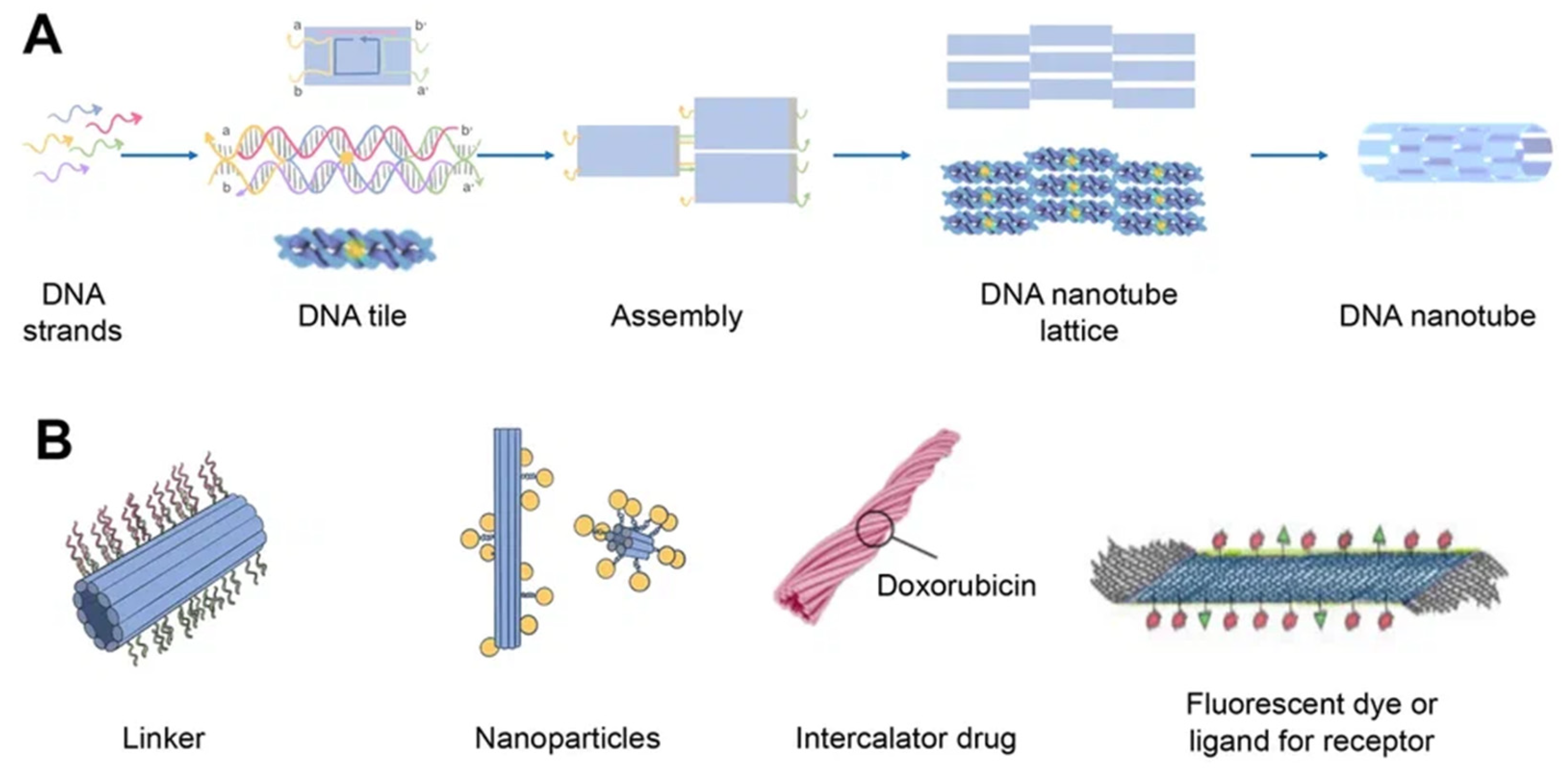
2.4. Aptamers
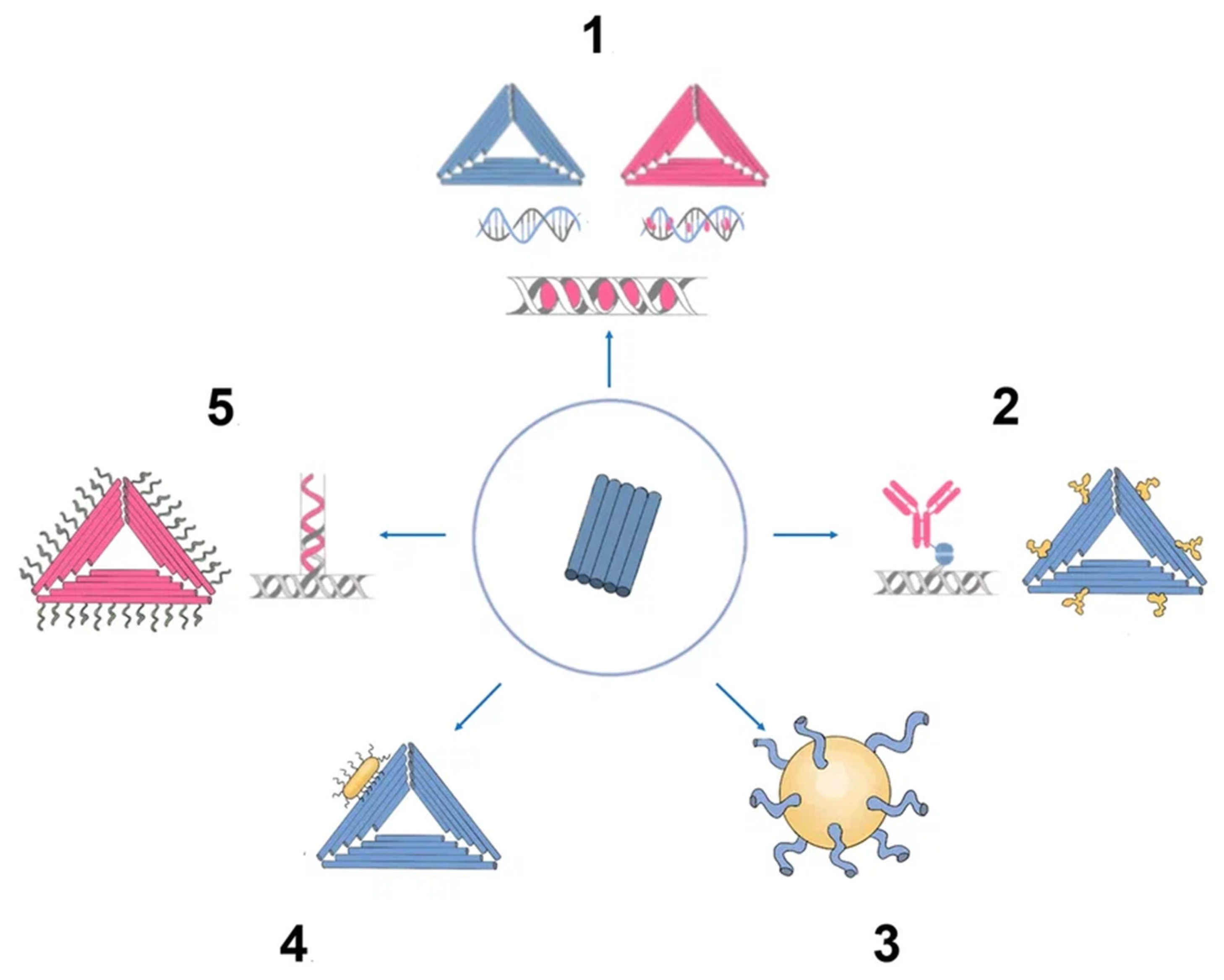
Aptamers are typically peptides or single-stranded DNA or RNA that are short in sequence and can bind to cellular targets with high selectivity. Secondary or tertiary structures of aptamers facilitate target binding and determine specificity and affinity [50][45]. Aptamers can be employed for targeting nanocarriers toward tissue overexpressing the target antigens [51][46]. Similar to DNA, aptamers can combine to generate complementary base pairs, which can then be used to build a variety of complex structures. They can be secondary structures such as the kissing hairpin, stem, loop, bugle, and pseudoknot. These secondary structures can then be combined to create certain three-dimensional structures, which the cell’s target molecule can subsequently recognize. The major forces that result in the creation of a three-dimensional structure and the attachment of an aptamer to a target include hydrogen bonds, van der Waals forces, and hydrophobic and electrostatic interactions. Similar to how antibodies bind to antigens, the creation of aptamer-target complexes is driven by a unique 3D interaction [52][47]. Systematic evolution of ligands by exponential enrichment (SELEX) selects nucleic acid aptamers from a library of random sequences, which then bind to the selected compounds with great specificity and affinity. Exponentially enriching a population of random sequence nucleic acid libraries enables the SELEX technique to evolve and select molecules with the highest affinities. SELEX is used for nucleic acids because of the possibility of conveniently amplifying affinity-selected molecules by RT–PCR or PCR. Aptamers can circumvent some of the disadvantages associated with the use of antibodies. For instance, aptamers are produced in vitro and can be chosen to target almost any protein, including toxins or nonimmunogenic proteins, in a relatively short amount of time, whereas the use of living animals is a drawback for antibody production [53][48]. Aptamers have received much attention recently in the field of biomedicine. Aptamers have characteristics similar to those of antibodies as well as particular benefits such as thermal stability, simplicity in synthesis, reversibility of target binding, and minimal immunogenicity. Chemical techniques make it simple to change aptamers by adding functional groups and/or lengthening them (Figure 5) [50][45]. Changes can confer nuclease resistance and prolong the circulation half-life. For instance, aptamers’ circulation half-life can be extended to many hours by adding carrier molecules to their ends, such as polyethylene glycol or cholesterol [54][49]. Moreover, aptamers might be conjugated to therapeutic compounds such as medications, carriers for drugs, poisons, or photosensitizers [53][48].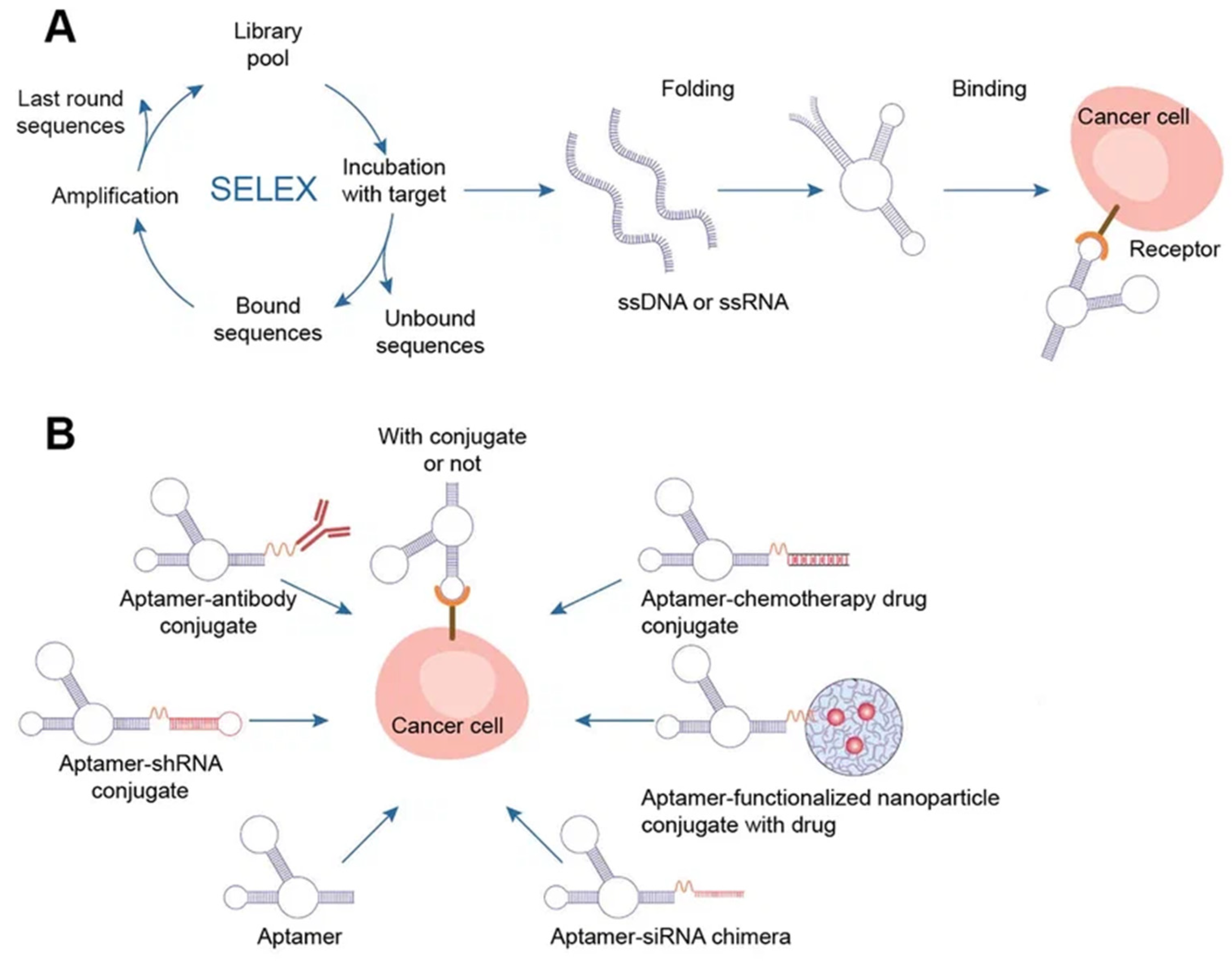
3. Delivery of DNA-Based Nanomaterials
3.1. Biodistribution and Biosafety of DNA-Based Nanomaterials
As of right now, the majority of oligonucleotide treatments (and nearly all licensed nucleic acid preparations) concentrate on either local delivery (for example, to the eyes or spinal cord) or hepatic delivery. As the liver is a strongly perfused organ, absorption of bigger nanoparticles and free oligonucleotides can happen quickly before renal clearance. Moreover, the liver has very high concentrations of receptors that might facilitate fast absorption and/or recycling. The development of effective methods for extrahepatic systemic distribution remains a key objective in the field of oligonucleotide treatment, despite the fact that other organs, such as the kidneys and spleen, are also locations of oligonucleotide accumulation [228,229][52][53]. Oligonucleotides are hydrophilic, negatively charged polymers that do not pass the plasma membrane by themselves [230][54]. Cellular DNAses are thought to break oligonucleotides within cells. On the other hand, the degradation process is linked to the uptake pathway of DNA nanostructures [13][55]. In addition, there are off-target interactions, toxicity depending on the sequence and chemical composition of oligonucleotides, and saturation of endogenous RNA processing pathways [231][56]. Data on the biodistribution of unmodified therapeutic DNA nanostructures, including aptamers, are limited. All unmodified oligonucleotides (including aptamers) have serious pharmacokinetic problems, including metabolic instability and rapid renal filtration without nonspecific protein binding [232][57]. For example, the half-lives of unmodified nucleotide aptamers in the blood are on the order of 2 min. Thus, short aptamer stability is a serious issue. Nucleic acid preparations must resist extracellular degradation [233][58], prevent the release of the drug bound to specific plasma proteins from circulation [234][59], and avoid removal by the reticuloendothelial system. Nucleic acid drug platforms must cross the capillary endothelium to the desired target, cross the plasma membrane, avoid lysosomal degradation [235][60], and be delivered to a specific target in the cell [230][54]. Therefore, the most commonly used strategies to improve drug delivery and safety from nucleic acids include chemical modification, covalent conjugation with cell-targeting or cell-penetrating molecules, and the use of nanoparticles to lessen adverse effects and boost the treatment efficiency of drug delivery technologies [230][54]. Most of the aptamers are within 50 nucleotides in length and fall within the renal filtration range. In the case of polymer conjugation, aptamers largely reduce renal filtration [236][61]. Additionally, chemical modifications are proposed to be incorporated into nucleotide sugars or internucleotide phosphodiester linkages to overcome this problem for aptamers and increase their serum half-life. In addition, mirror L-DNA can solve the problem of low stability of natural DNA in blood serum, which affects the pharmacokinetics and biological distribution [237][62]. To date, many DNA-based carriers have been shown to have little cytotoxicity and are mainly used as a hub for integrating drug loading, targeting, and release modules together [238][63]. There is currently very limited information on the safety and efficacy of treatments using DNA nanostructures. Some experiments have been carried out on cell culture, as well as several animal studies [239][64]. One of the biggest challenges for TDNs in further clinical use is whether TDNs can cause unpredictable gene recombination or deleterious effects in the liver or kidneys. Nevertheless, it was shown that TDN has better biological safety, lower biotoxicity, and higher transport efficiency than other DNA carriers in cell culture [240][65]. For example, the biosafety of TDN for DOX delivery has been shown [15][10]. Zhang et al. demonstrated that triangular origami DNA loaded with doxorubicin could be an effective and safe innovation platform for the treatment of breast cancer in nude mice [241][66]. Recently, the safety of DNA origami at physiological pH for noncancerous cells and its cytotoxicity for cancer cells at the pH of solid tumors have been demonstrated [242][67]. In general, aptamers have also been shown to have little or no side effects and are safe. Aptamers did not lead to the activation of the immune system [243][68]. Many analyses in preclinical and early clinical trials have not shown complement activation. Additionally, no off-target side effects were identified [244][69]. In 2022, the first phase I human clinical trial of the ApTOLL DNA aptamer was conducted and the results in healthy male volunteers demonstrated great safety and a suitable pharmacokinetic profile of aptamer. The infusion’s termination resulted in the highest concentration, and the mean half-life was 9.3 h. At any dose or with any researched form of administration, serious adverse effects or biochemical abnormalities were not seen. However, the study showed no accumulation of ApTOLL [55][50]. However, the shelf life of DNA nanostructures can raise safety concerns. For instance, because DNA sequences in nanostructures are complicated and unpredictable, foreign DNA may at random interact with cellular RNA and result in other possible chronic toxicities. Moreover, the immunological response to DNA nanostructures in vivo has not been well studied. Immunostimulatory activity through a TLR9-independent mechanism should not be disregarded, despite the fact that numerous research claim that CpG-free DNA nanostructures have little immunogenicity [245][70].3.2. Cellular Uptake of DNA Nanostructures
Endocytosis is the process through which macromolecules, their complexes, and large particles enter the cell. Endocytosis is classified into two types: phagocytosis, which consumes particles larger than 250 nm, and pinocytosis, which consumes fluid and particles that phagocytosis does not consume. Pinocytosis occurs in all cell types via four different mechanisms: macropinocytosis, clathrin-mediated endocytosis, caveolar-mediated endocytosis, and clathrin- and caveolin-independent endocytosis [246][71]. Other pathways, with the exception of macropinocytosis, are directly controlled by cargo molecule activity. Cargo molecules bind to receptors and form receptor–ligand complexes attracting certain effectors to specific parts of the plasma membrane [247][72]. Depending on their size and the proteins involved, oligonucleotides enter cells through a variety of endocytosis processes (Figure 6). Despite the fact that all endocytosis routes lead to the creation of endosomes, molecules entering via distinct channels may end up at different downstream locations. The majority of absorbed oligonucleotides end up in late endosomes and lysosomes. There is, nevertheless, some partial translocation to other membrane-bound compartments. Oligonucleotides inside the endomembrane compartment are pharmacologically inactive, and only a tiny percentage of internalized oligonucleotides may reach the cytosol and nucleus on their own. The method of injection may influence oligonucleotide cellular uptake [248][73].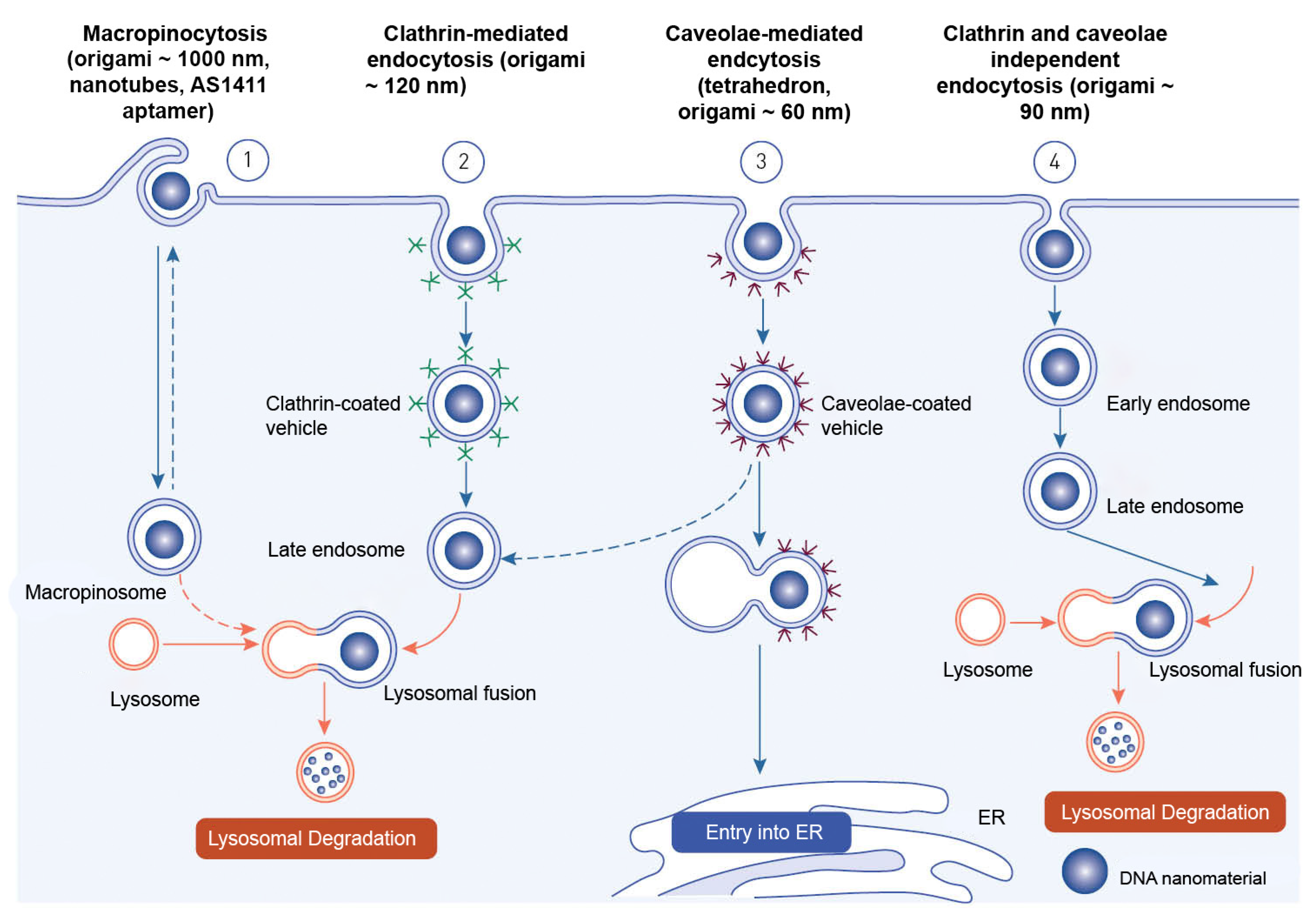
References
- Zhang, T.; Tian, T.; Zhou, R.; Li, S.; Ma, W.; Zhang, Y.; Liu, N.; Shi, S.; Li, Q.; Xie, X.; et al. Design, Fabrication and Applications of Tetrahedral DNA Nanostructure-Based Multifunctional Complexes in Drug Delivery and Biomedical Treatment. Nat. Protoc. 2020, 15, 2728–2757.
- Zhang, Y.; Tu, J.; Wang, D.; Zhu, H.; Maity, S.K.; Qu, X.; Bogaert, B.; Pei, H.; Zhang, H. Programmable and Multifunctional DNA-Based Materials for Biomedical Applications. Adv. Mater. 2018, 30, 1703658.
- Xia, K.; Kong, H.; Cui, Y.; Ren, N.; Li, Q.; Ma, J.; Cui, R.; Zhang, Y.; Shi, J.; Li, Q.; et al. Systematic Study in Mammalian Cells Showing No Adverse Response to Tetrahedral DNA Nanostructure. ACS Appl. Mater. Interfaces 2018, 10, 15442–15448.
- Pei, H.; Zuo, X.; Zhu, D.; Huang, Q.; Fan, C. Functional DNA Nanostructures for Theranostic Applications. Acc. Chem. Res. 2014, 47, 550–559.
- Liang, L.; Li, J.; Li, Q.; Huang, Q.; Shi, J.; Yan, H.; Fan, C. Single-Particle Tracking and Modulation of Cell Entry Pathways of a Tetrahedral DNA Nanostructure in Live Cells. Angew. Chem. Int. Ed. 2014, 53, 7745–7750.
- Bagalkot, V.; Farokhzad, O.C.; Langer, R.; Jon, S. An Aptamer–Doxorubicin Physical Conjugate as a Novel Targeted Drug-Delivery Platform. Angew. Chem. Int. Ed. 2006, 45, 8149–8152.
- Erben, C.M.; Goodman, R.P.; Turberfield, A.J. Single-Molecule Protein Encapsulation in a Rigid DNA Cage. Angew. Chem. Int. Ed. 2006, 45, 7414–7417.
- Ma, W.; Zhan, Y.; Zhang, Y.; Mao, C.; Xie, X.; Lin, Y. The Biological Applications of DNA Nanomaterials: Current Challenges and Future Directions. Signal Transduct. Target. Ther. 2021, 6, 351.
- Duangrat, R.; Udomprasert, A.; Kangsamaksin, T. Tetrahedral DNA Nanostructures as Drug Delivery and Bioimaging Platforms in Cancer Therapy. Cancer Sci. 2020, 111, 3164–3173.
- Zhang, G.; Zhang, Z.; Yang, J. DNA Tetrahedron Delivery Enhances Doxorubicin-Induced Apoptosis of HT-29 Colon Cancer Cells. Nanoscale Res. Lett. 2017, 12, 495.
- Wiraja, C.; Zhu, Y.; Lio, D.C.S.; Yeo, D.C.; Xie, M.; Fang, W.; Li, Q.; Zheng, M.; Van Steensel, M.; Wang, L.; et al. Framework Nucleic Acids as Programmable Carrier for Transdermal Drug Delivery. Nat. Commun. 2019, 10, 1147.
- Yan, J.; Zhan, X.; Zhang, Z.; Chen, K.; Wang, M.; Sun, Y.; He, B.; Liang, Y. Tetrahedral DNA Nanostructures for Effective Treatment of Cancer: Advances and Prospects. J. Nanobiotechnology 2021, 19, 412.
- Xie, X.; Shao, X.; Ma, W.; Zhao, D.; Shi, S.; Li, Q.; Lin, Y. Overcoming Drug-Resistant Lung Cancer by Paclitaxel Loaded Tetrahedral DNA Nanostructures. Nanoscale 2018, 10, 5457–5465.
- Kim, K.-R.; Kim, D.-R.; Lee, T.; Yhee, J.Y.; Kim, B.-S.; Kwon, I.C.; Ahn, D.-R. Drug Delivery by a Self-Assembled DNA Tetrahedron for Overcoming Drug Resistance in Breast Cancer Cells. Chem. Commun. 2013, 49, 2010.
- Kim, K.; Lee, H.-B.-R.; Johnson, R.W.; Tanskanen, J.T.; Liu, N.; Kim, M.-G.; Pang, C.; Ahn, C.; Bent, S.F.; Bao, Z. Selective Metal Deposition at Graphene Line Defects by Atomic Layer Deposition. Nat. Commun. 2014, 5, 4781.
- Tang, W.; Xu, H.; Kopelman, R.; Philbert, M.A. Photodynamic Characterization and In Vitro Application of Methylene Blue-Containing Nanoparticle Platforms¶. Photochem. Photobiol. 2007, 81, 242–249.
- Yousuf, S.; Enoch, I.V.M.V. Binding Interactions of Naringenin and Naringin with Calf Thymus DNA and the Role of β-Cyclodextrin in the Binding. AAPS PharmSciTech 2013, 14, 770–781.
- Kim, K.-R.; Bang, D.; Ahn, D.-R. Nano-Formulation of a Photosensitizer Using a DNA Tetrahedron and Its Potential for in Vivo Photodynamic Therapy. Biomater. Sci. 2016, 4, 605–609.
- Kim, K.-R.; Hwang, D.; Kim, J.; Lee, C.-Y.; Lee, W.; Yoon, D.S.; Shin, D.; Min, S.-J.; Kwon, I.C.; Chung, H.S.; et al. Streptavidin-Mirror DNA Tetrahedron Hybrid as a Platform for Intracellular and Tumor Delivery of Enzymes. J. Control. Release 2018, 280, 1–10.
- Tian, Y.; Huang, Y.; Gao, P.; Chen, T. Nucleus-Targeted DNA Tetrahedron as a Nanocarrier of Metal Complexes for Enhanced Glioma Therapy. Chem. Commun. 2018, 54, 9394–9397.
- Yang, J.; Jiang, Q.; He, L.; Zhan, P.; Liu, Q.; Liu, S.; Fu, M.; Liu, J.; Li, C.; Ding, B. Self-Assembled Double-Bundle DNA Tetrahedron for Efficient Antisense Delivery. ACS Appl. Mater. Interfaces 2018, 10, 23693–23699.
- Zhang, C.; Han, M.; Zhang, F.; Yang, X.; Du, J.; Zhang, H.; Li, W.; Chen, S. Enhancing Antitumor Efficacy of Nucleoside Analog 5-Fluorodeoxyuridine on HER2-Overexpressing Breast Cancer by Affibody-Engineered DNA Nanoparticle. Int. J. Nanomed. 2020, 15, 885–900.
- Rothemund, P.W.K. Folding DNA to Create Nanoscale Shapes and Patterns. Nature 2006, 440, 297–302.
- Strauss, M.T.; Schueder, F.; Haas, D.; Nickels, P.C.; Jungmann, R. Quantifying Absolute Addressability in DNA Origami with Molecular Resolution. Nat. Commun. 2018, 9, 1600.
- Kearney, C.J.; Lucas, C.R.; O’Brien, F.J.; Castro, C.E. DNA Origami: Folded DNA-Nanodevices That Can Direct and Interpret Cell Behavior. Adv. Mater. 2016, 28, 5509–5524.
- Dey, S.; Fan, C.; Gothelf, K.V.; Li, J.; Lin, C.; Liu, L.; Liu, N.; Nijenhuis, M.A.D.; Saccà, B.; Simmel, F.C.; et al. DNA Origami. Nat. Rev. Methods Prim. 2021, 1, 13.
- Babatunde, B.; Arias, D.S.; Cagan, J.; Taylor, R.E. Generating DNA Origami Nanostructures through Shape Annealing. Appl. Sci. 2021, 11, 2950.
- Chen, T.; Ren, L.; Liu, X.; Zhou, M.; Li, L.; Xu, J.; Zhu, X. DNA Nanotechnology for Cancer Diagnosis and Therapy. Int. J. Mol. Sci. 2018, 19, 1671.
- Weiden, J.; Bastings, M.M.C. DNA Origami Nanostructures for Controlled Therapeutic Drug Delivery. Curr. Opin. Colloid Interface Sci. 2021, 52, 101411.
- Jiang, Q.; Song, C.; Nangreave, J.; Liu, X.; Lin, L.; Qiu, D.; Wang, Z.-G.; Zou, G.; Liang, X.; Yan, H.; et al. DNA Origami as a Carrier for Circumvention of Drug Resistance. J. Am. Chem. Soc. 2012, 134, 13396–13403.
- Majikes, J.M.; Nash, J.A.; LaBean, T.H. Search for Effective Chemical Quenching to Arrest Molecular Assembly and Directly Monitor DNA Nanostructure Formation. Nanoscale 2017, 9, 1637–1644.
- Halley, P.D.; Lucas, C.R.; McWilliams, E.M.; Webber, M.J.; Patton, R.A.; Kural, C.; Lucas, D.M.; Byrd, J.C.; Castro, C.E. Daunorubicin-Loaded DNA Origami Nanostructures Circumvent Drug-Resistance Mechanisms in a Leukemia Model. Small 2016, 12, 308–320.
- Zhuang, X.; Ma, X.; Xue, X.; Jiang, Q.; Song, L.; Dai, L.; Zhang, C.; Jin, S.; Yang, K.; Ding, B.; et al. A Photosensitizer-Loaded DNA Origami Nanosystem for Photodynamic Therapy. ACS Nano 2016, 10, 3486–3495.
- Chandrasekaran, A.R.; Anderson, N.; Kizer, M.; Halvorsen, K.; Wang, X. Beyond the Fold: Emerging Biological Applications of DNA Origami. ChemBioChem 2016, 17, 1081–1089.
- O’Neill, P.; Rothemund, P.W.K.; Kumar, A.; Fygenson, D.K. Sturdier DNA Nanotubes via Ligation. Nano Lett. 2006, 6, 1379–1383.
- Sun, L.; Yu, L.; Shen, W. DNA Nanotechnology and Its Applications in Biomedical Research. J. Biomed. Nanotechnol. 2014, 10, 2350–2370.
- Liu, X.; Zhao, Y.; Liu, P.; Wang, L.; Lin, J.; Fan, C. Biomimetic DNA Nanotubes: Nanoscale Channel Design and Applications. Angew. Chem. Int. Ed. 2019, 58, 8996–9011.
- Aldaye, F.A.; Lo, P.K.; Karam, P.; McLaughlin, C.K.; Cosa, G.; Sleiman, H.F. Modular Construction of DNA Nanotubes of Tunable Geometry and Single- or Double-Stranded Character. Nat. Nanotechnol. 2009, 4, 349–352.
- Burns, J.R. Introducing Bacteria and Synthetic Biomolecules along Engineered DNA Fibers. Small 2021, 17, 2100136.
- Liang, L.; Shen, J.-W.; Wang, Q. Molecular Dynamics Study on DNA Nanotubes as Drug Delivery Vehicle for Anticancer Drugs. Colloids Surf. B Biointerfaces 2017, 153, 168–173.
- Henry, S.J.W.; Stephanopoulos, N. Functionalizing DNA Nanostructures for Therapeutic Applications. WIREs Nanomed. Nanobiotechnol. 2021, 13, e1729.
- Ko, S.; Liu, H.; Chen, Y.; Mao, C. DNA Nanotubes as Combinatorial Vehicles for Cellular Delivery. Biomacromolecules 2008, 9, 3039–3043.
- Kocabey, S.; Ekim Kocabey, A.; Schneiter, R.; Rüegg, C. Membrane-Interacting DNA Nanotubes Induce Cancer Cell Death. Nanomaterials 2021, 11, 2003.
- Li, C.H.; Lv, W.Y.; Yan, Y.; Yang, F.F.; Zhen, S.J.; Huang, C.Z. Nucleolin-Targeted DNA Nanotube for Precise Cancer Therapy through Förster Resonance Energy Transfer-Indicated Telomerase Responsiveness. Anal. Chem. 2021, 93, 3526–3534.
- Kong, H.Y.; Byun, J. Nucleic Acid Aptamers: New Methods for Selection, Stabilization, and Application in Biomedical Science. Biomol. Ther. 2013, 21, 423–434.
- Dutta, B.; Barick, K.C.; Hassan, P.A. Recent Advances in Active Targeting of Nanomaterials for Anticancer Drug Delivery. Adv. Colloid Interface Sci. 2021, 296, 102509.
- Zhou, J.; Rossi, J. Aptamers as Targeted Therapeutics: Current Potential and Challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202.
- Santosh, B.; Yadava, P.K. Nucleic Acid Aptamers: Research Tools in Disease Diagnostics and Therapeutics. Biomed Res. Int. 2014, 2014, 540451.
- Nimjee, S.M.; White, R.R.; Becker, R.C.; Sullenger, B.A. Aptamers as Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 61–79.
- Hernández-Jiménez, M.; Martín-Vílchez, S.; Ochoa, D.; Mejía-Abril, G.; Román, M.; Camargo-Mamani, P.; Luquero-Bueno, S.; Jilma, B.; Moro, M.A.; Fernández, G.; et al. First-in-Human Phase I Clinical Trial of a TLR4-Binding DNA Aptamer, ApTOLL: Safety and Pharmacokinetics in Healthy Volunteers. Mol. Ther.-Nucleic Acids 2022, 28, 124–135.
- Liu, S.; Xu, Y.; Jiang, X.; Tan, H.; Ying, B. Translation of Aptamers toward Clinical Diagnosis and Commercialization. Biosens. Bioelectron. 2022, 208, 114168.
- Jackson, A.L.; Bartz, S.R.; Schelter, J.; Kobayashi, S.V.; Burchard, J.; Mao, M.; Li, B.; Cavet, G.; Linsley, P.S. Expression Profiling Reveals Off-Target Gene Regulation by RNAi. Nat. Biotechnol. 2003, 21, 635–637.
- Doench, J.G.; Petersen, C.P.; Sharp, P.A. SiRNAs Can Function as MiRNAs. Genes Dev. 2003, 17, 438–442.
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in Oligonucleotide Drug Delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694.
- Hu, Q.; Li, H.; Wang, L.; Gu, H.; Fan, C. DNA Nanotechnology-Enabled Drug Delivery Systems. Chem. Rev. 2019, 119, 6459–6506.
- Grimm, D.; Streetz, K.L.; Jopling, C.L.; Storm, T.A.; Pandey, K.; Davis, C.R.; Marion, P.; Salazar, F.; Kay, M.A. Fatality in Mice Due to Oversaturation of Cellular MicroRNA/Short Hairpin RNA Pathways. Nature 2006, 441, 537–541.
- Kovacevic, K.D.; Gilbert, J.C.; Jilma, B. Pharmacokinetics, Pharmacodynamics and Safety of Aptamers. Adv. Drug Deliv. Rev. 2018, 134, 36–50.
- Geary, R.S.; Norris, D.; Yu, R.; Bennett, C.F. Pharmacokinetics, Biodistribution and Cell Uptake of Antisense Oligonucleotides. Adv. Drug Deliv. Rev. 2015, 87, 46–51.
- Shemesh, C.S.; Yu, R.Z.; Gaus, H.J.; Seth, P.P.; Swayze, E.E.; Bennett, F.C.; Geary, R.S.; Henry, S.P.; Wang, Y. Pharmacokinetic and Pharmacodynamic Investigations of ION-353382, a Model Antisense Oligonucleotide: Using Alpha-2-Macroglobulin and Murinoglobulin Double-Knockout Mice. Nucleic Acid Ther. 2016, 26, 223–235.
- Sahay, G.; Querbes, W.; Alabi, C.; Eltoukhy, A.; Sarkar, S.; Zurenko, C.; Karagiannis, E.; Love, K.; Chen, D.; Zoncu, R.; et al. Efficiency of SiRNA Delivery by Lipid Nanoparticles Is Limited by Endocytic Recycling. Nat. Biotechnol. 2013, 31, 653–658.
- Ashrafuzzaman, M. Aptamers as Both Drugs and Drug-Carriers. Biomed Res. Int. 2014, 2014, 697923.
- Kim, K.-R.; Kim, H.Y.; Lee, Y.-D.; Ha, J.S.; Kang, J.H.; Jeong, H.; Bang, D.; Ko, Y.T.; Kim, S.; Lee, H.; et al. Self-Assembled Mirror DNA Nanostructures for Tumor-Specific Delivery of Anticancer Drugs. J. Control. Release 2016, 243, 121–131.
- Jiang, S.; Ge, Z.; Mou, S.; Yan, H.; Fan, C. Designer DNA Nanostructures for Therapeutics. Chem 2021, 7, 1156–1179.
- Coleridge, E.L.; Dunn, K.E. Assessing the Cost-Effectiveness of DNA Origami Nanostructures for Targeted Delivery of Anti-Cancer Drugs to Tumours. Biomed. Phys. Eng. Express 2020, 6, 065030.
- Li, S.; Tian, T.; Zhang, T.; Cai, X.; Lin, Y. Advances in Biological Applications of Self-Assembled DNA Tetrahedral Nanostructures. Mater. Today 2019, 24, 57–68.
- Zhang, Q.; Jiang, Q.; Li, N.; Dai, L.; Liu, Q.; Song, L.; Wang, J.; Li, Y.; Tian, J.; Ding, B.; et al. DNA Origami as an In Vivo Drug Delivery Vehicle for Cancer Therapy. ACS Nano 2014, 8, 6633–6643.
- Wang, Y.; Benson, E.; Fördős, F.; Lolaico, M.; Baars, I.; Fang, T.; Teixeira, A.I.; Högberg, B. DNA Origami Penetration in Cell Spheroid Tissue Models Is Enhanced by Wireframe Design. Adv. Mater. 2021, 33, 2008457.
- Bouchard, P.R.; Hutabarat, R.M.; Thompson, K.M. Discovery and Development of Therapeutic Aptamers. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 237–257.
- Szebeni, J. Complement Activation-Related Pseudoallergy: A Stress Reaction in Blood Triggered by Nanomedicines and Biologicals. Mol. Immunol. 2014, 61, 163–173.
- Liu, X.; Xu, Y.; Yu, T.; Clifford, C.; Liu, Y.; Yan, H.; Chang, Y. A DNA Nanostructure Platform for Directed Assembly of Synthetic Vaccines. Nano Lett. 2012, 12, 4254–4259.
- Park, J.H.; Oh, N. Endocytosis and Exocytosis of Nanoparticles in Mammalian Cells. Int. J. Nanomedicine 2014, 9 (Suppl. S1), 51–63.
- Wan, L.-Y.; Yuan, W.-F.; Ai, W.-B.; Ai, Y.-W.; Wang, J.-J.; Chu, L.-Y.; Zhang, Y.-Q.; Wu, J.-F. An Exploration of Aptamer Internalization Mechanisms and Their Applications in Drug Delivery. Expert Opin. Drug Deliv. 2019, 16, 207–218.
- Juliano, R.L. The Delivery of Therapeutic Oligonucleotides. Nucleic Acids Res. 2016, 44, 6518–6548.
- Alamudi, S.H.; Kimoto, M.; Hirao, I. Uptake Mechanisms of Cell-Internalizing Nucleic Acid Aptamers for Applications as Pharmacological Agents. RSC Med. Chem. 2021, 12, 1640–1649.
- El-Sayed, A.; Harashima, H. Endocytosis of Gene Delivery Vectors: From Clathrin-Dependent to Lipid Raft-Mediated Endocytosis. Mol. Ther. 2013, 21, 1118–1130.
- Shapira, A.; Livney, Y.D.; Broxterman, H.J.; Assaraf, Y.G. Nanomedicine for Targeted Cancer Therapy: Towards the Overcoming of Drug Resistance. Drug Resist. Updat. 2011, 14, 150–163.
