Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Zeinab Ezzeddine and Version 2 by Camila Xu.
The pathogenic anaerobic bacteria Yersinia pestis (Y. pestis), which is well known as the plague causative agent, has the ability to escape or inhibit innate immune system responses, which can result in host death even before the activation of adaptive responses. Yersinia pestis produces two metallophores: yersiniabactin, for iron chelation (siderophore), and an opine type metallophore called yersinopine.
- Yersinia pestis
- pathogenesis
- metallophores
- metal ions
1. Introduction
The Gram-negative pathogen Yersinia pestis (Y. pestis), which is responsible for causing bubonic, pneumonic, and septicemic plague among humans, emerged from the enteropathogen Yersinia pseudotuberculosis [1]. This pathogen is considered a zoonosis because it is carried by rodents worldwide and transmitted within them through a flea vector (Xenopsylla cheopis) [2][3][2,3]. Effective inter-human transmission may occur through aerosols leading to the development of pneumonic plague [4][5][4,5]. However, recent evidence shows that inter-human transmission via aerosol is apparently insignificant [6] and may be due to body lice and fleas [1].
Y. pestis evolved multiple factors that enable it to colonize its hosts which include both mammals and insects [7]. An infected flea causes bubonic plague through its bite. Once deposited on the dermis, the bacterium spreads and colonizes the lymph nodes, leading to ‘‘bubo’’ which is the inflammation of the lymph nodes [8].
2. Yersiniabactin
The transition metal iron is crucial for almost all pathogenic bacteria including Y. pestis. In the host metal ions’ scarce environment, bacteria developed various strategies to sequester iron. One of these strategies is the synthesis and secretion of siderophores which are low molecular weight metabolites specific for obtaining ferric ions. The highly pathogenic Y. pestis produces the siderophore yersiniabactin (Ybt), which has shown an important role in the acquisition of iron and murine pathogenicity [9][42]. Moreover, it was demonstrated that Y. pestis strains with mutations in genes responsible for Ybt biosynthesis or uptake caused an almost complete inability to generate fatal bubonic and pneumonic plague [10][11][43,44]. These findings prove that Ybt is associated with virulence besides acquiring iron.2.1. Yersiniabactin Biosynthesis
The siderophore Ybt is composed of thiazoline rings that are responsible for Fe3+ binding with a formation constant of 4 × 1036 [12][45]. The synthesis of Ybt is carried on by NRPS (nonribosomal peptide synthetase) mechanism combined with PKS (polyketide synthase). Ybt is constructed from salicylate, a linker malonyl group, three cysteines, and three methyl groups from S-adenosylmethionine [13][46]. These molecules yield a siderphore whose structure is composed of four rings (one salicylate, two thiazolines, and one thiazolidine) as shown in Figure 1.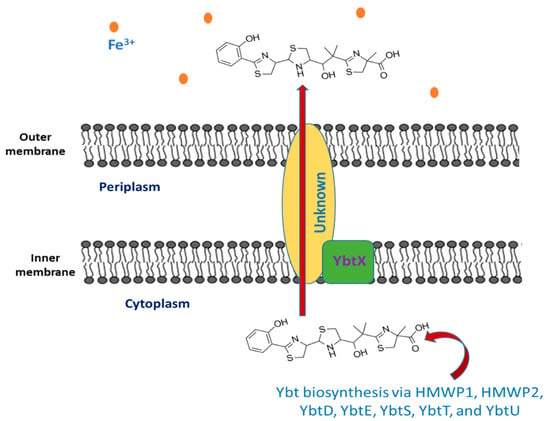
Figure 1.
The export of yersiniabactin.
2.2. The Export of Ybt
Once the synthesis of Ybt is completed, it is exported outside the bacterial cell, but the mechanism underlying this process is still unknown. Y. pestis comprise YbtX, which is an inner membrane protein assigned to be a part of the Ybt efflux system because it resembles both AlcS and EntS exporters in alcaligin and enterobactin siderophores in Bordetella and E. coli respectively [18][19][20][51,52,53]. However, experiments showed that a Y. pestis ybtX mutant can secrete Ybt, uptake iron-loaded Ybt, and can grow under iron-scarce conditions as well [19][52]. The YbtX function is not fully understood, although it is involved in the secretion of Ybt, it is not essential in the process.2.3. Iron Chelation by Ybt
As mentioned previously, Ybt contains a benzene ring, another thiazolidine ring, and two thiazoline rings. This structure provides five chiral centers (donor positions), enabling it to form a stable complex with trivalent ferric iron cations (Fe3+) with 1:1 complexes. The Ybt potential donor groups are: both the nitrogen found in the first thiazoline ring and the phenolic hydroxy group, either the nitrogen or the sulfur present in the thiazolidine moiety along with the aliphatic hydroxy group, as well as the carboxy group and the nitrogen of the terminal thiazoline ring [21][54].2.4. The Intake of Iron-Loaded Ybt
After Ybt chelates iron, the bacterium uptakes the loaded siderophore. The first step includes binding to the TonB-dependent receptor Psn, which is followed by outer membrane translocation. In Gram-negative bacteria, the function of TonB is associated with ExbB and ExbD, yet it is not ascertained in Y. pestis. Then, the iron-loaded Ybt is transported from the periplasm into the cytoplasm via the YbtP-YbtQ ABC transporter. The latter are similar inner membrane proteins with both ATPase and permease domains and are usually components of Type I secretion systems [22][23][55,56]. In the cytoplasm, Fe is then released in the cytoplasm from the siderophore by a reduction in ferric ions to ferrous ions or by Ybt degradation. The process of Ybt-Fe3+ uptake is demonstrated in Figure 2. Genes that encode either type of iron release mechanisms are not found within the high pathogenicity island (HPI) that is responsible for encoding most ybt genes. Noteworthily, Y. pestis mutant strains in either psn or tonB demonstrated both growth and iron uptake defects. Moreover, despite the presence of a second TonB-like gene (hasB), it is unable to be an alternative to TonB in taking iron through the Ybt system [24][25][57,58]. Moreover, strains with ybtP or ybtQ mutations have a similar phenotype psn mutant and showed reduced iron uptake and growth but without a notable reduction in Ybt production [22][55].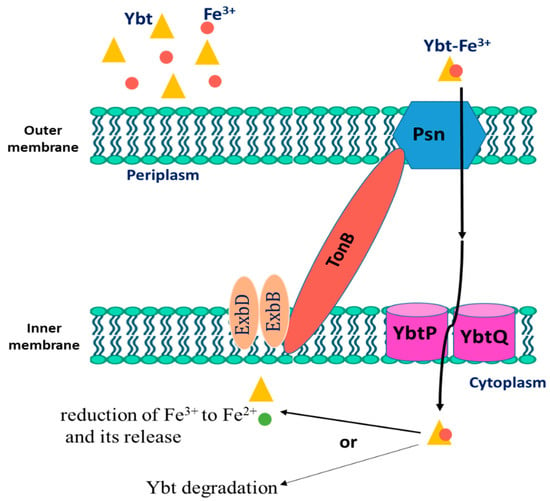
Figure 2.
The uptake process of Fe
3+
-loaded Yersiniabactin (Ybt) by
Y. pestis
.
2.5. Genetic Regulation of Yersiniabactin (Ybt)
In Y. pestis, the ybt locus which is composed of four operons is responsible for Ybt biosynthesis, uptake, and possibly export. Out of the four operons, two are monocistronic, where one of them contains five genes encoding the biosynthetic enzymes, while the fourth operon genes encode the proteins involved in Ybt uptake and biosynthesis. Except for ybtD, fur, and the transport components (tonB, exbB, and exbC), all other genes involved in the function or regulation or function of the Ybt system are encoded within this locus [26][59]. The ybt locus is found within the high pathogenicity island (HPI) that was identified, for the first time, in the three Yersinia species causing pathogenesis in mammals. The Y. pestis HPI is found within the pigmentation (pgm) locus and comprises approximately one-third of it. The latter is an unstable wide area of the Y. pestis chromosome consisting of an HPI segment linked to a pigmentation segment [27][60]. The HPI is distributed widely among the Enterobacteriaceae family members. The IS100 related to HPI of Y. pestis is not found in the other Enterobacteriaceae. The majority of the organisms having the HPI produce the proteins HMWP1 and HMWP2 in iron-scarce conditions and synthesize the siderophore Ybt [26][59]. As in other Gram-negative bacteria, Y. pestis produces iron uptake regulation protein (Fur), which is responsible for repressing the transcription of the promoters associated with a Fur binding sequence (FBS) once iron is in excess [27][60]. All of the four ybt operons that lie within the HPI possess FBSs promoters and are repressed via Fur approximately 12-fold (psn), 8-fold (irp2-irp1-ybtUTE), 11-fold (ybtA), and 55-fold (ybtPQXS) upon the growth with 10 μM iron compared to the growth in deferrated, defined media (PMH/PMH2) without additional iron [24][25][26][27][28][57,58,59,60,61]. ybtD, on the contrary, that is encoded outside the HPI and pgm locus is not Fur regulated.2.6. The Role of Yersiniabactin in Overcoming Zinc Restriction
It was recently demonstrated that Ybt binds. thus enhancing the growth Y. pestis lacking ZnuABC transporter in zinc scarce medium. These findings suggest that Ybt may have a role in overcoming zinc nutritional immunity in addition to its role in iron sequestering. To test this assumption, Price et al. used an iron-mediated nutritional immunity defective mouse and demonstrated the contribution of Ybt to virulence independent of iron. In addition, they found that Y. pestis utilizes Ybt to compete for zinc with calprotectin. Furthermore, they discovered that in flea midgut, this pathogen relies on Ybt to survive zinc limitation [29][62]. Older studies showed that the irp2 gene of Y. pestis, responsible for HMWP2 encoding, is needed for its growth in zinc-deficient medium for strains lacking ZnuABC [30][39]. Additionally, they showed that YbtX is required as well for the uptake of Ybt-dependent Zn2+ while both Psn and TonB, along with YbtPQ, are not required for zinc acquisition [31][63]. These results were further proved by Bobrov et al. who demonstrated that strains lacking both ybtX and znu genes were avirulent for bubonic and pneumonic plague in mice models at the time the virulence ability was conserved in strains mutant only for ybtX [32][64]. All these findings prove that Ybt has a role in the mechanism of zinc acquisition in Y. pestis, enabling it to overcome zinc limitation upon infecting both insects and mammals.3. Yersinopine
Plants produce the metabolite nicotianamine, which is used in metal ions homeostasis such as zinc, iron, and nickel [33][65]. It is also synthesized by filamentous fungi [34][66] along with some mosses [35][67]. On the other hand, it was found that bacteria produce opine-type metallophores resembling nicotianamine, [36][68] in particular Staphylococcus aureus, Pseudomonas aeruginosa, and Yersinia pestis. These bacteria produce, respectively, staphylopine, pseudopaline, and yersinopine containing an imidazole ring and three carboxylic groups [37][38][69,70]. The structure of yersinopine is shown in Figure 3.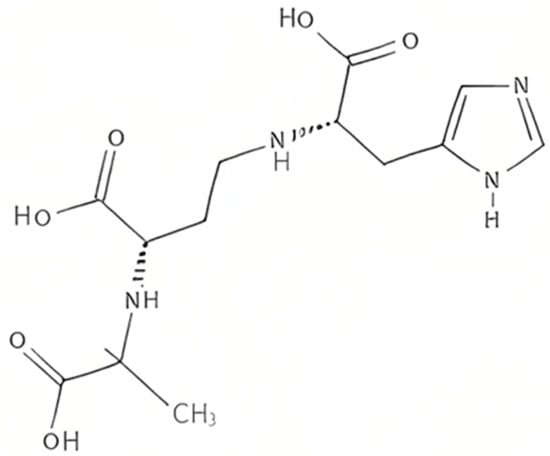
Figure 3.
The structure of yersinopine.
