DNA damage in astronauts induced by cosmic radiation poses a major barrier to human space exploration. Cellular responses and repair of the most lethal DNA double-strand breaks (DSBs) are crucial for genomic integrity and cell survival. Post-translational modifications (PTMs), including phosphorylation, ubiquitylation, and SUMOylation, are among the regulatory factors modulating a delicate balance and choice between predominant DSB repair pathways, such as non-homologous end joining (NHEJ) and homologous recombination (HR). RIn this researchersview, we focused on the engagement of proteins in the DNA damage response (DDR) modulated by phosphorylation and ubiquitylation, including ATM, DNA-PKcs, CtIP, MDM2, and ubiquitin ligases. The involvement and function of acetylation, methylation, PARylation, and their essential proteins were also investigated, providing a repository of candidate targets for DDR regulators. However, there is a lack of radioprotectors in spite of their consideration in the discovery of radiosensitizers. We proposed new perspectives for the research and development of future agents against space radiation by the systematic integration and utilization of evolutionary strategies, including multi-omics analyses, rational computing methods, drug repositioning, and combinations of drugs and targets, which may facilitate the use of radioprotectors in practical applications in human space exploration to combat fatal radiation hazards.
- radiation protection
- space radiation
- DSB
- post-translational modification
- drug target
- drug discovery
1. Introduction
2. PTMs in the Choice of DNA Repair Pathways
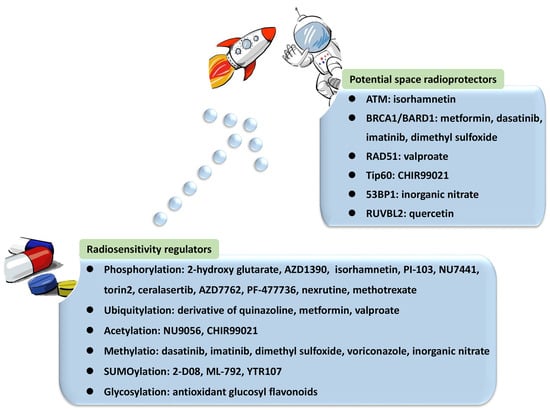
HR is a critical pathway for the error-free repair of DNA DSBs, while NHEJ always occurs in the absence of a sister chromatid, leading to error-prone repair and more mutations [42][43][44][42,43,44]. NHEJ was reported to be the predominant DNA repair pathway in mammalian cells [45]. The choice of the repair pathway was found to be tightly associated with the cell cycle, as NHEJ is the default repair pathway [46] usually executed in the G1 phase of the cell cycle in a rapid and high-capacity manner [42][47][42,47]. Unlike NHEJ, which may occur throughout the entire cell cycle, HR is largely limited to the S/G2 phases [42][48][42,48] and is conducted more slowly than NHEJ [47]. The underlying mechanism is that DSB repair is executed with higher efficiency during the S phase. DSB processing and checkpoint activation are much more efficient in the G2/M phase than in the G1 phase [49]. In general, the 5′-3′ degradation of DSB ends is needed for the loading of checkpoint and recombination proteins in all HR reactions [50][51][50,51]. The generation of long 3′ single-strand DNA (ssDNA) overhangs mediated by DNA helicases and exonuclease in DNA end resection was proven to be an essential committed process in HR [47][48][52][53][54][47,48,52,53,54]. In contrast, NHEJ requires DNA ends that have not been resected instead of 3′ ssDNA tails. DNA end resection is not needed, leading to the joining of two DNA ends with few references to the DNA sequence [47][48][55][56][47,48,55,56]. Therefore, controlling DNA end resection is one of the processes affecting whether DNA repair is conducted by NHEJ or HR [35][40][35,40]. For example, the 53BP1-RIF1-shieldin complex cooperates with the CTC1-STN1-TEN1 (CST)/Pol α-Prim complex in regulating the generation of 3′ overhangs, which are essential for DNA end protection and switching the DSB repair mode to NHEJ [53][57][53,57]. In contrast, BRCA1 promotes HR and antagonizes NHEJ by stimulating end resection [58][59][58,59]. Several key proteins and their complexes play regulatory roles in NHEJ. For instance, the Ku70-Ku80 heterodimer is central in initiating NHEJ by recognizing DSB ends and recruiting DNA-PKcs to DSB sites [51][60][61][62][63][51,60,61,62,63]. In addition, 53BP1 stimulates NHEJ by recruiting other DDR proteins such as ATM and inhibiting DNA end resection processing by protecting broken DNA ends with its co-factors PTIP, RIF1-shieldin, or REV7/MAD2L2 [48][56][61][62][64][65][66][67][48,56,61,62,64,65,66,67]. In contrast, the important factors in HR mainly include BRCA1/2, EXO1, MRE11 [47][48][64][68][69][47,48,64,68,69], and RAD51 and its paralogs [43][47][69][70][43,47,69,70]. Among them, BRCA1 directly affects the DSB repair pathway choice by regulating the initiation of end resection [52][59][52,59]. The preservation of long-term resection activity requires EXO1 exonuclease [71], the deficiency of which contributes to the accumulation of unprocessed DSBs and HR failure [72]. MRE11 exonuclease activity is needed for the assembly of a series of proteins to DSB sites to mediate extended-end resection for HR [73]. HR is orchestrated by several PTMs with elaborate primary mechanisms. The first one is phosphorylation. Switching the meiotic recombination mode of HR was reported to occur by the phosphorylation of RAD54 and HED1, downregulating RAD51 activity by suppressing Rad51/Rad54 complex formation [74][75][74,75]. Secondly, SUMOylation is important in HR. It affects all steps in HR and exerts various regulatory functions on substrates [76]. Evidence indicated that SUMOylation induced by topoisomerase 1-binding arginine/serine-rich protein (TOPORS) was essential for the recruitment of RAD51 to the damaged sites and the support of HR repair, maintaining genomic stability [77]. On the other hand, NHEJ might be associated with phosphorylation and methylation by DNA-PKcs and 53BP1, respectively. NHEJ and HR are competitive, and their balance is finely modulated by bioprocesses that include PTMs. Ubiquitination is the most vital PTM, playing a specific role in the recruitment and enrichment of DDR factors at DSB sites in chromosomes and governing DNA repair pathway choices between NHEJ and HR. DDR proteins are mainly assembled by ubiquitin E3 ligases RNF8 and RNF168, followed by accurate repair processes [35]. The ubiquitylation-dependent DSB repair pathway choice is frequently associated with DNA end resection. For example, Cullin3-KLHL15 ubiquitin ligase participates in CtIP protein turnover through the ubiquitin–proteasome pathway, fine-tuning DNA end resection and impacting the balance between HR and NHEJ [78]. RING domain-containing E3 ligase RNF138 is involved in the ubiquitination of Ku80 during the S phase and its removal from DSB sites, stimulating DSB end resection and promoting HR initiation [79]. In addition to DNA end resection, ubiquitylation also modulates the choice of DNA repair pathways by altering the expression of specific DDR proteins. CtIP, which is a target of anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase, is downregulated during G1 and G2 phases and reduces HR [80]. CtIP ubiquitylation and upregulation are stimulated by UBE2Ds and RNF138 at DNA damage sites, promoting DNA repair by HR [81]. Moreover, ubiquitination affects the DNA repair pathway choice by regulating histone H2A at Lys15 (H2AK15ub) and initiating downstream signaling events [82]. Phosphorylation is another major PTM involved in the balance of DNA repair pathways. The phosphorylation of ubiquitin at Thr12 (pUbT12) influences DDR by regulating the activity of 53BP1 in damaged chromosomes [83], and 53BP1 inhibits excessive DNA end resection and promotes repair by NHEJ through different phosphoprotein interactions [84]. RIF1 is prominent at DSB sites in the G1 phase of the cell cycle by the ATM-associated phosphorylation of 53BP1, ensuring the dominant position of NHEJ in this phase [58][59][85][58,59,85]. Collectively, a variety of proteins and their complexes were revealed to act in the complicated response mechanisms to DNA lesions induced by IR, participating in distinct PTMs and coordinating NHEJ, HR, and their balance in DNA repair (Figure 1). These essential factors could be properly categorized and investigated from the view of PTMs, including phosphorylation, ubiquitylation, acetylation, and methylation. Compounds targeting these factors influence DNA repair after IR, leading to radiosensitization or radioprotective effects (Table 1). Some of them have been approved for regulating DDR, and more candidates are under development. They provide resources in the discovery of future space radioprotectors (Figure 2).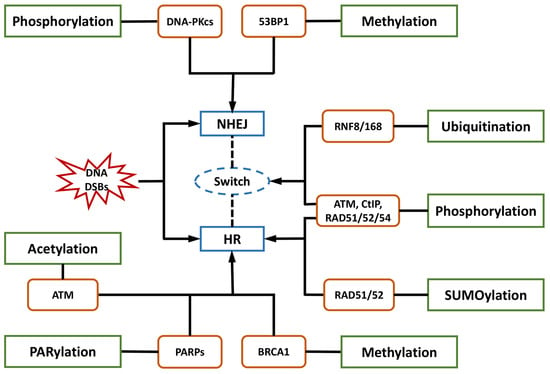
| Post-Translational Modification (PTM) | Essential Factor | Description | Application | Reference |
|---|---|---|---|---|
| Phosphorylation | ATM |
|
|
[86][87][88][89][90][91][92][93][86,87,88,89,90,91,92,93] |
| DNA-PKcs |
|
|
[88][94][95][96][88,94,95,96] | |
| CtIP |
|
|
[42][97][98][99][42,97,98,99] | |
| ATR |
|
|
[100][101][100,101] | |
| CHK1 |
|
|
[101][102][103][101,102,103] | |
| RAD51/52/54 |
|
|
[43][70][104[107][43,70][,104105][,105106],106,107] | |
| H2AX |
|
/ | [108][109][108,109] | |
| Ubiquitylation | RNF8 |
|
|
[110][111][112][110,111,112] |
| RNF168 |
|
|
[47][110][111][113][114][115][116][47,110,111,113,114,115,116] | |
| REV1 |
|
|
[117][118][119][120][117,118,119,120] | |
| MDM2 |
|
|
[89][121][122][123][124][89,121,122,123,124] | |
| BRCA1/BARD1 |
|
|
[83][125][126][127][83,125,126,127] | |
| RAD51 |
|
|
[128] | |
| Acetylation | Tip60 |
|
|
[90][129][130][90[,129131,130],131] |
| Methylation | BRCA1 |
|
|
[132][133][134][135][132,133,134,135] |
| 53BP1 |
|
|
[136][136[137],137] | |
| PARylation | PARP1 |
|
|
[138][139][138,139] |
| Neddylation | NEDD8 |
|
|
[140][141][142][143][140,141,142,143] |
| SUMOylation | / |
|
|
[144] |
| MEIIL3 |
|
|
[145] | |
| NPM1 |
|
|
[146] | |
| Glycosylation | OGT |
|
|
[147][148][147,148] |
| NEIL3 |
|
|
[149][150][151][152][149,150,151,152] | |
| RUVBL1/2 |
|
|
[150][153][150,153] | |
| Kcr | CDYL1 |
|
/ | [154] |
| CBP/P300 |
|
|
[155][156][157][158][155,156,157,158] |
