Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Jessie Wu and Version 3 by Jessie Wu.
Prostate cancer (PCa) is the most common urological malignancy and brings great health threats to men.
- PCa
- cancer stem cells
- signaling pathway
- tumor
1. Introduction
Cancer cell heterogeneity can result from the differentiation program driven by a subset of unique cancer cells called cancer stem cells (CSCs), which have self-renewal potential and can differentiate into various types of cells in a symmetrical or asymmetrical cell divided manner [1][2][3]. In 2005, with the successful isolation of CD44+/α2β1hi/CD133+ enriched cells from a human prostate tumor, Collins et al. found that this small population of prostate cancer (PCa) cells possessed self-renewal ability and were enabled to differentiate into non-clonal tumor cells [4]. Other studies also demonstrated that CD44+ and PSA-/lo PCa cells shared similar properties with CD44+/α2β1hi/CD133+ enriched cells [5][6][7]. With decades of research, scientists have gradually recognized the existence and significance of these fractional cells in PCa using various biomarkers (Table 1) [8][9][10][11][12][13][14][15][16][17][18][19][20][21][22][23][24][25][26][27][28][29][30][31][32], which have stem cell characteristics and are then referred as prostate cancer stem cells (PCSCs). Now, it is clear that PCSCs are involved in the initiation, progression and therapeutic resistance of PCa.
Table 1. Biomarkers for prostate cancer stem cells.
| Marker | Function | Reference |
|---|---|---|
| CD44 | CD44 is a glycoprotein involved in cell migration, adhesion and signal transduction | Kalantari E, et al., 2017 [8] |
| CD133 | CD133 is a transmembrane glycoprotein involved in cell self-renewal, differentiation and tumor invasion | Kalantari E, et al., 2017 [8] |
| CD117 | CD117 is a transmembrane glycoprotein involved in cell self-renewal, differentiation and tumor invasion | Harris KS, et al., 2021 [9] |
| α2β1 | α2β1 is associated with tumor invasion and proliferation | Aldahish A, et al., 2019 [10] Naci D, et al., 2015 [11] |
| EpCAM | EpCAM is a calcium independent adhesion molecule between epithelial cells involved in the process of epithelial cell carcinogenesis | Witte KE, et al., 2021 [12] |
| SOX2 | SOX2 plays an important role in maintaining stem cell pluripotency and self-renewal | Lee Y, et al., 2021 [13] Vaddi PK, et al., 2019 [14] |
| EZH2 | EZH2 is closely related to cell migration and invasion, tumor development and stem cell self-renewal | Huang J, et al., 2021 [15] |
| CXCR4 | CXCR4 is a specific receptor involved in physiological mechanisms such as HIV-1, hematopoiesis, embryonic development and tumor migration | Li Y, et al., 2019 [16] Chatterjee S, et al., 2014 [17] |
| TRA-1-60 | A cell surface epitope of human embryonic, embryonal germline and teratocarcinoma stem cells | Schafer C, et al., 2020 [18] |
| CD151 | CD151 is associated with tumor initiation, metastasis, and angiogenesis | Wong AH, et al., 2020 [19] |
| OCT-3/4 | OCT-3/4 is an essential transcription factor that maintains the multidirectional differentiation potential of embryonic stem cells and primordial germ cells | Wang X, et al., 2021 [20] Fujimura T, et al., 2014 [21] |
| Smo | Smo is a transmembrane protein that mediates Hedgehog signaling to the intracellular compartment | Lou H, et al., 2020 [22] |
| Nanog | Nanog is a transcription factor with an important role in stem cell self-renewal and maintenance of pluripotency | Verma S, et al., 2020 [23] Liu C, et al., 2020 [24] |
| Bmi-1 | Bmi-1 is associated with maintenance of self-renewal of prostate stem cells and inhibition of PTEN in PCa | Li Y, et al., 2017 [25] Yoo YA, et al., 2016 [26] |
| TWIST | TWIST is a transcription factor with a helix-loop-helix structure and associated with tumor invasion and metastasis | Lee Y, et al., 2021 [13] |
| CD24 | CD24 is a cell adhesion molecule involved in the regulation of B-cell proliferation and maturation | Costa CD, et al.,2019 [27] |
| CD166 | CD166 is a leukocyte adhesion factor associated with cell adhesion and tumor metastasis | Wei GJ, et al., 2019 [28] van Lersner A, et al., 2019 [29] |
| CD49b | CD49b is also called integrin α2, a cell surface receptor associated with adhesion and lymphocyte activation | Bansal N, et al., 2016 [30] Erb HHH, et al., 2018 [31] |
| ABCG-2 | ABCG-2 contributes to the resistance to chemotherapeutic drugs | Kim WT, et al., 2017 [32] |
2. The Role of Prostate Cancer Stem Cells in Prostate Cancer Initiation
A number of literature studies have highlighted cancer stem cells as the origin of tumor initiation, including PCa. In the prostate gland, a small population of prostate stem cells (PSCs) undergo an oncogenic transformation to PCSCs when the external environment is unfavorable (Figure 1). Then, the omnipotent PCSCs give rise to various sub-types of prostate cancer cells, either AR-dependent or -independent, forming PCa heterogeneity. It seems that luminal-like PCa originates from progenitor cells in luminal epithelial cells. However, α2β1hi/CD133+ prostate stem cells were previously identified to be located in the basal layer [33]. Moreover, Witte et al. impressively found that it was the basal epithelial cells, but not luminal epithelial cells, growing to the prostate gland structure when they were subcutaneously implanted into nonobese diabetic scid gamma (NSG) mice [34]. Another study revealed that only basal epithelial cells with oncogenic induction led to prostatic intraepithelial neoplasia (PIN) and PCa in NSG mice [35]. All these findings together suggest that PCSCs in basal epithelial cells are the origin of PCa.
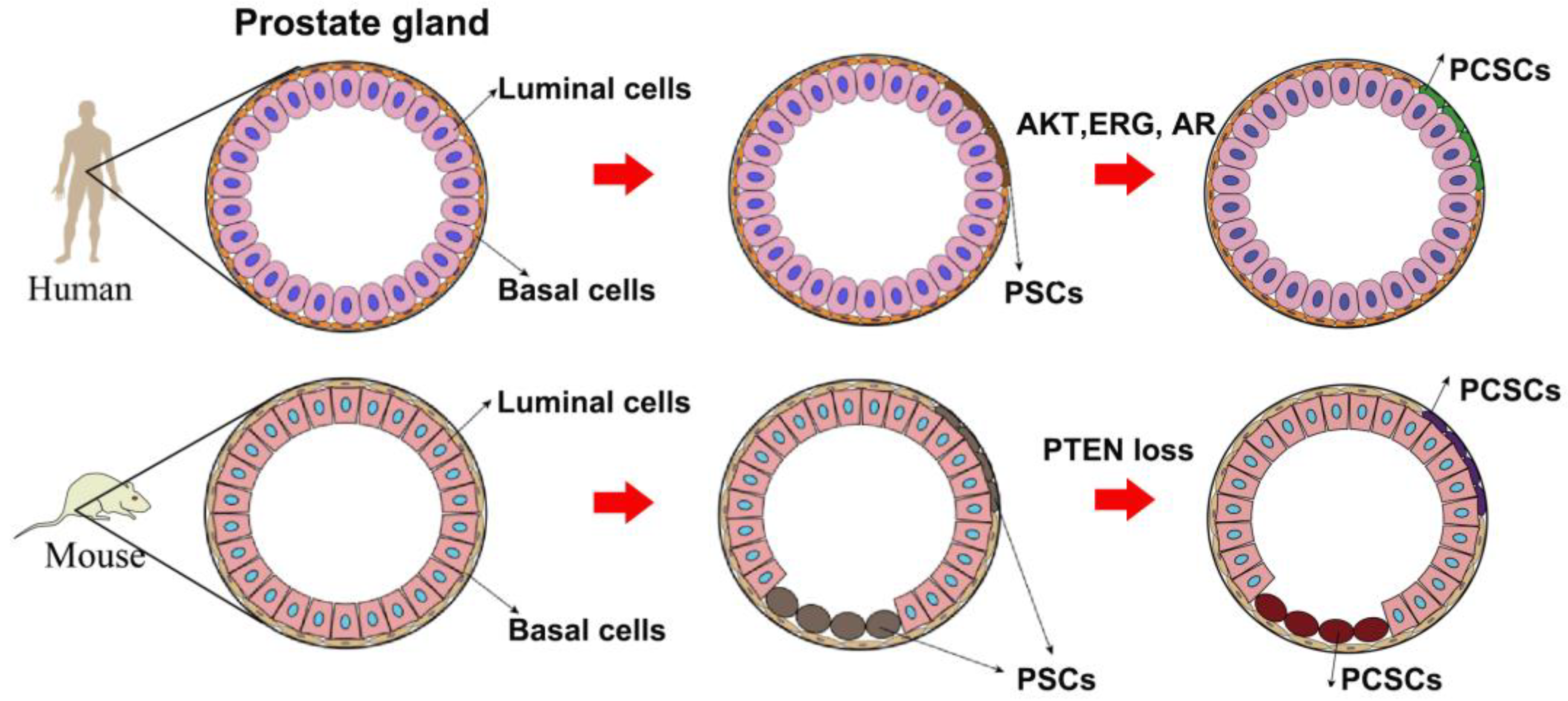
Figure 1. A model of the origin of prostate cancer stem cells.
However, the origin of PCa in mice is different from that in humans. Studies by lineage tracing system elucidated that PSCs can be found in both the luminal and basal layers [36][37]. Further investigation illuminated that phosphatase and tensin homolog (PTEN) loss in either the basal layer or luminal layer led to PCa development. In the view of a CSCs-based hypothesis, PSCs in either the luminal layer or basal layer will transform to PCSCs when suffering from genetic alteration such as PTEN loss [38]. Although there is a little discrepancy between mouse PCa and human PCa regarding their origin (Figure 1), the concept of PCSCs as PCa origin has been widely acknowledged by many scientists: PSCs in the prostate epithelial cells undergo genomic instability to give rise to PCSCs when the environment is unfavorable, finally leading to PCa development.
3. The Role of Prostate Cancer Stem Cells in Prostate Cancer Progression and Anti-Androgen Resistance
PCa with 2–3 years androgen deprivation therapy (ADT) treatment will eventually develop to castration-resistant disease, a lethal stage owing to its metastatic potential. By bypassing this systemic androgen suppression, AR signaling hijacks several means in order to efficiently respond to the castrated androgen level. AR amplification, AR mutation and selective up-regulation of AR co-regulators are the more frequently observed biological processes in castration-resistant prostate cancer (CRPC) as compared with primary tumors [39][40][41][42].
Growing evidence also illustrates that ADT treatment expands the population of PCSCs [43][44][45], which have no response to androgenic sources. Continuous AR signaling inhibition of CRPC is fulfilled by the treatment of FDA approved anti-androgen enzalutamide [46][47], which competes androgen to bind AR and suppresses AR nuclear imports [48]. However, it only benefits CRPC by extending to a five month survival [49]. One popular mechanism responsible for the acquired enzalutamide resistance is the selective expression of truncated AR variants. For example, ARv7, which lacks an androgen responsive domain, is a crucially causal factor controlling enzalutamide resistance [50][51][52]. Another important mechanism responsible for anti-androgen resistance is lineage plasticity [53], which is possibly driven by PCSCs. Accumulative evidence suggests that AR-targeted therapies including ADT and anti-androgen treatment considerably enrich the population of PCSCs by facilitating the transition of AR-dependent PCa cells to AR-independent cells with stem-cell-like states or multi-lineage stages [54][55]. PCSCs then re-differentiate into AR-therapy-resistant sub-clones such as neuroendocrine PCa cells or other lineage sub-clones inherently resistant to enzalutamide treatment. In this regard, PCSCs play critical roles in PCa progression and anti-androgen resistance.
References
- Yadav, A.K.; Desai, N.S. Cancer Stem Cells: Acquisition, Characteristics, Therapeutic Implications, Targeting Strategies and Future Prospects. Stem Cell Rev. Rep. 2019, 15, 331–355.
- Barbato, L.; Bocchetti, M.; Di Biase, A.; Regad, T. Cancer Stem Cells and Targeting Strategies. Cells 2019, 8, 926.
- Lytle, N.K.; Barber, A.G.; Reya, T. Stem cell fate in cancer growth, progression and therapy resistance. Nat. Rev. Cancer 2018, 18, 669–680.
- Collins, A.T.; Berry, P.A.; Hyde, C.; Stower, M.J.; Maitland, N.J. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005, 65, 10946–10951.
- Chen, X.; Li, Q.; Liu, X.; Liu, C.; Liu, R.; Rycaj, K.; Zhang, D.; Liu, B.; Jeter, C.; Calhoun-Davis, T.; et al. Defining a Population of Stem-like Human Prostate Cancer Cells That Can Generate and Propagate Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2016, 22, 4505–4516.
- Acikgoz, E.; Soner, B.C.; Ozdil, B.; Guven, M. CD133+/CD44+ prostate cancer stem cells exhibit embryo-like behavior patterns. Acta Histochem. 2021, 123, 151743.
- Binal, Z.; Acikgoz, E.; Kizilay, F.; Oktem, G.; Altay, B. Cross-talk between ribosome biogenesis, translation, and mTOR in CD133+ 4/CD44+ prostate cancer stem cells. Clin. Transl. Oncol. 2020, 22, 1040–1048.
- Kalantari, E.; Asgari, M.; Nikpanah, S.; Salarieh, N.; Asadi Lari, M.H.; Madjd, Z. Co-Expression of Putative Cancer Stem Cell Markers CD44 and CD133 in Prostate Carcinomas. Pathol. Oncol. Res. 2017, 23, 793–802.
- Harris, K.S.; Shi, L.; Foster, B.M.; Mobley, M.E.; Elliott, P.L.; Song, C.J.; Watabe, K.; Langefeld, C.D.; Kerr, B.A. CD117/c-kit defines a prostate CSC-like subpopulation driving progression and TKI resistance. Sci. Rep. 2021, 11, 1465.
- Aldahish, A.; Kale, A.; Aljameeli, A.; Shah, G.V. Calcitonin induces stem cell-like phenotype in prostate cancer cells. Endocr. Relat. Cancer 2019, 26, 815–828.
- Naci, D.; Vuori, K.; Aoudjit, F. Alpha2beta1 integrin in cancer development and chemoresistance. Semin. Cancer Biol. 2015, 35, 145–153.
- Witte, K.E.; Pfitzenmaier, J.; Storm, J.; Lutkemeyer, M.; Wimmer, C.; Schulten, W.; Czaniera, N.; Geisler, M.; Forster, C.; Wilkens, L.; et al. Analysis of Several Pathways for Efficient Killing of Prostate Cancer Stem Cells: A Central Role of NF-kappaB RELA. Int. J. Mol. Sci. 2021, 22, 8901.
- Lee, Y.; Yoon, J.; Ko, D.; Yu, M.; Lee, S.; Kim, S. TMPRSS4 promotes cancer stem-like properties in prostate cancer cells through upregulation of SOX2 by SLUG and TWIST1. J. Exp. Clin. Cancer Res. 2021, 40, 372.
- Vaddi, P.K.; Stamnes, M.A.; Cao, H.; Chen, S. Elimination of SOX2/OCT4-Associated Prostate Cancer Stem Cells Blocks Tumor Development and Enhances Therapeutic Response. Cancers 2019, 11, 1331.
- Huang, J.; Xu, Y.; Wang, F.; Wang, H.; Li, L.; Deng, Y.; Cai, L. Long Noncoding RNA SPRY4-IT1 Modulates Ketamine-Induced Neurotoxicity in Human Embryonic Stem Cell-Derived Neurons through EZH2. Dev. Neurosci. 2021, 43, 9–17.
- Li, Y.; He, Y.; Butler, W.; Xu, L.; Chang, Y.; Lei, K.; Zhang, H.; Zhou, Y.; Gao, A.C.; Zhang, Q.; et al. Targeting cellular heterogeneity with CXCR2 blockade for the treatment of therapy-resistant prostate cancer. Sci. Transl. Med. 2019, 11, eaax0428.
- Chatterjee, S.; Behnam Azad, B.; Nimmagadda, S. The intricate role of CXCR4 in cancer. Adv. Cancer Res. 2014, 124, 31–82.
- Schafer, C.; Ju, Y.; Tak, Y.; Vazquez, C.; Han, S.J.; Tan, E.; Shay, J.W.; Holmqvist, M.; Danuser, G.; Schopperle, W.M.; et al. TRA-1-60-positive/CD45(low) cells found in the peripheral blood of prostate cancer patients with metastatic disease—A proof-of-concept study. Heliyon 2020, 6, e03263.
- Wong, A.H.; Tran, T. CD151 in Respiratory Diseases. Front. Cell Dev. Biol. 2020, 8, 64.
- Wang, X.; Yang, J.Y.; Cai, J.; Zhang, D.J.; Zhao, L.; Luo, L.H.; Xiong, Y.; Zhang, T.; Jin, M. MiR-543/Numb promotes proliferation, metastasis, and stem-like cell traits of prostate cancer cells. Am. J. Transl. Res. 2021, 13, 617–631.
- Fujimura, T.; Takahashi, S.; Urano, T.; Takayama, K.; Sugihara, T.; Obinata, D.; Yamada, Y.; Kumagai, J.; Kume, H.; Ouchi, Y.; et al. Expression of androgen and estrogen signaling components and stem cell markers to predict cancer progression and cancer-specific survival in patients with metastatic prostate cancer. Clin. Cancer Res. 2014, 20, 4625–4635.
- Lou, H.; Li, H.; Huehn, A.R.; Tarasova, N.I.; Saleh, B.; Anderson, S.K.; Dean, M. Genetic and Epigenetic Regulation of the Smoothened Gene (SMO) in Cancer Cells. Cancers 2020, 12, 2219.
- Verma, S.; Shankar, E.; Kalayci, F.N.C.; Mukunda, A.; Alassfar, M.; Singh, V.; Chan, E.R.; MacLennan, G.T.; Gupta, S. Androgen Deprivation Induces Transcriptional Reprogramming in Prostate Cancer Cells to Develop Stem Cell-Like Characteristics. Int. J. Mol. Sci. 2020, 21, 9568.
- Liu, C.; Sheng, M.; Lin, L.; Li, H.; Guo, S.; Zhang, J.; Chen, G.; Chen, H. NANOG regulates the proliferation of PCSCs via the TGF-beta1/SMAD pathway. Open Med. 2020, 15, 841–849.
- Li, Y.; Wang, L.; Liu, J.; Zhang, P.; An, M.; Han, C.; Li, Y.; Guan, X.; Zhang, K. O-GlcNAcylation modulates Bmi-1 protein stability and potential oncogenic function in prostate cancer. Oncogene 2017, 36, 6293–6305.
- Yoo, Y.A.; Roh, M.; Naseem, A.F.; Lysy, B.; Desouki, M.M.; Unno, K.; Abdulkadir, S.A. Bmi1 marks distinct castration-resistant luminal progenitor cells competent for prostate regeneration and tumour initiation. Nat. Commun. 2016, 7, 12943.
- Costa, C.D.; Justo, A.A.; Kobayashi, P.E.; Story, M.M.; Palmieri, C.; Laufer Amorim, R.; Fonseca-Alves, C.E. Characterization of OCT3/4, Nestin, NANOG, CD44 and CD24 as stem cell markers in canine prostate cancer. Int. J. Biochem. Cell Biol. 2019, 108, 21–28.
- Wei, G.J.; Chao, Y.H.; Tung, Y.C.; Wu, T.Y.; Su, Z.Y. A Tangeretin Derivative Inhibits the Growth of Human Prostate Cancer LNCaP Cells by Epigenetically Restoring p21 Gene Expression and Inhibiting Cancer Stem-like Cell Proliferation. AAPS J. 2019, 21, 86.
- von Lersner, A.; Droesen, L.; Zijlstra, A. Modulation of cell adhesion and migration through regulation of the immunoglobulin superfamily member ALCAM/CD166. Clin. Exp. Metastasis 2019, 36, 87–95.
- Bansal, N.; Bartucci, M.; Yusuff, S.; Davis, S.; Flaherty, K.; Huselid, E.; Patrizii, M.; Jones, D.; Cao, L.; Sydorenko, N.; et al. BMI-1 Targeting Interferes with Patient-Derived Tumor-Initiating Cell Survival and Tumor Growth in Prostate Cancer. Clin. Cancer Res. 2016, 22, 6176–6191.
- Erb, H.H.H.; Guggenberger, F.; Santer, F.R.; Culig, Z. Interleukin-4 induces a CD44(high) /CD49b(high) PC3 subpopulation with tumor-initiating characteristics. J. Cell. Biochem. 2018, 119, 4103–4112.
- Kim, W.T.; Ryu, C.J. Cancer stem cell surface markers on normal stem cells. BMB Rep. 2017, 50, 285–298.
- Williamson, S.C.; Hepburn, A.C.; Wilson, L.; Coffey, K.; Ryan-Munden, C.A.; Pal, D.; Leung, H.Y.; Robson, C.N.; Heer, R. Human alpha(2)beta(1)(HI) CD133(+VE) epithelial prostate stem cells express low levels of active androgen receptor. PLoS ONE 2012, 7, e48944.
- Lawson, D.A.; Xin, L.; Lukacs, R.U.; Cheng, D.; Witte, O.N. Isolation and functional characterization of murine prostate stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 181–186.
- Goldstein, A.S.; Huang, J.; Guo, C.; Garraway, I.P.; Witte, O.N. Identification of a cell of origin for human prostate cancer. Science 2010, 329, 568–571.
- Choi, N.; Zhang, B.; Zhang, L.; Ittmann, M.; Xin, L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell 2012, 21, 253–265.
- Wang, Z.A.; Mitrofanova, A.; Bergren, S.K.; Abate-Shen, C.; Cardiff, R.D.; Califano, A.; Shen, M.M. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat. Cell Biol. 2013, 15, 274–283.
- Wang, Z.A.; Toivanen, R.; Bergren, S.K.; Chambon, P.; Shen, M.M. Luminal cells are favored as the cell of origin for prostate cancer. Cell Rep. 2014, 8, 1339–1346.
- Aurilio, G.; Cimadamore, A.; Mazzucchelli, R.; Lopez-Beltran, A.; Verri, E.; Scarpelli, M.; Massari, F.; Cheng, L.; Santoni, M.; Montironi, R. Androgen Receptor Signaling Pathway in Prostate Cancer: From Genetics to Clinical Applications. Cells 2020, 9, 2653.
- Kohli, M.; Ho, Y.; Hillman, D.W.; Van Etten, J.L.; Henzler, C.; Yang, R.; Sperger, J.M.; Li, Y.; Tseng, E.; Hon, T.; et al. Androgen Receptor Variant AR-V9 Is Coexpressed with AR-V7 in Prostate Cancer Metastases and Predicts Abiraterone Resistance. Clin. Cancer Res. 2017, 23, 4704–4715.
- Korpal, M.; Korn, J.M.; Gao, X.; Rakiec, D.P.; Ruddy, D.A.; Doshi, S.; Yuan, J.; Kovats, S.G.; Kim, S.; Cooke, V.G.; et al. An F876L mutation in androgen receptor confers genetic and phenotypic resistance to MDV3100 (enzalutamide). Cancer Discov. 2013, 3, 1030–1043.
- Liu, G.; Sprenger, C.; Sun, S.; Epilepsia, K.S.; Haugk, K.; Zhang, X.; Coleman, I.; Nelson, P.S.; Plymate, S. AR variant ARv567es induces carcinogenesis in a novel transgenic mouse model of prostate cancer. Neoplasia 2013, 15, 1009–1017.
- Kushwaha, P.P.; Verma, S.; Kumar, S.; Gupta, S. Role of prostate cancer stem-like cells in the development of antiandrogen resistance. Cancer Drug Resist. 2022, 5, 459–471.
- Horning, A.M.; Wang, Y.; Lin, C.K.; Louie, A.D.; Jadhav, R.R.; Hung, C.N.; Wang, C.M.; Lin, C.L.; Kirma, N.B.; Liss, M.A.; et al. Single-Cell RNA-seq Reveals a Subpopulation of Prostate Cancer Cells with Enhanced Cell-Cycle-Related Transcription and Attenuated Androgen Response. Cancer Res. 2018, 78, 853–864.
- Sanchez, B.G.; Bort, A.; Vara-Ciruelos, D.; Diaz-Laviada, I. Androgen Deprivation Induces Reprogramming of Prostate Cancer Cells to Stem-Like Cells. Cells 2020, 9, 1441.
- Davis, I.D.; Martin, A.J.; Stockler, M.R.; Begbie, S.; Chi, K.N.; Chowdhury, S.; Coskinas, X.; Frydenberg, M.; Hague, W.E.; Horvath, L.G.; et al. Enzalutamide with Standard First-Line Therapy in Metastatic Prostate Cancer. N. Engl. J. Med. 2019, 381, 121–131.
- Beer, T.M.; Armstrong, A.J.; Rathkopf, D.E.; Loriot, Y.; Sternberg, C.N.; Higano, C.S.; Iversen, P.; Bhattacharya, S.; Carles, J.; Chowdhury, S.; et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N. Engl. J. Med. 2014, 371, 424–433.
- Wu, Z.; Wang, K.; Yang, Z.; Pascal, L.E.; Nelson, J.B.; Takubo, K.; Wipf, P.; Wang, Z. A novel androgen receptor antagonist JJ-450 inhibits enzalutamide-resistant mutant AR(F876L) nuclear import and function. Prostate 2020, 80, 319–328.
- Denmeade, S.R.; Wang, H.; Agarwal, N.; Smith, D.C.; Schweizer, M.T.; Stein, M.N.; Assikis, V.; Twardowski, P.W.; Flaig, T.W.; Szmulewitz, R.Z.; et al. TRANSFORMER: A Randomized Phase II Study Comparing Bipolar Androgen Therapy Versus Enzalutamide in Asymptomatic Men with Castration-Resistant Metastatic Prostate Cancer. J. Clin. Oncol. 2021, 39, 1371–1382.
- Antonarakis, E.S.; Lu, C.; Wang, H.; Luber, B.; Nakazawa, M.; Roeser, J.C.; Chen, Y.; Mohammad, T.A.; Chen, Y.; Fedor, H.L.; et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014, 371, 1028–1038.
- Scher, H.I.; Lu, D.; Schreiber, N.A.; Louw, J.; Graf, R.P.; Vargas, H.A.; Johnson, A.; Jendrisak, A.; Bambury, R.; Danila, D.; et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker with Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol. 2016, 2, 1441–1449.
- Wang, R.; Sun, Y.; Li, L.; Niu, Y.; Lin, W.; Lin, C.; Antonarakis, E.S.; Luo, J.; Yeh, S.; Chang, C. Preclinical Study using Malat1 Small Interfering RNA or Androgen Receptor Splicing Variant 7 Degradation Enhancer ASC-J9((R)) to Suppress Enzalutamide-resistant Prostate Cancer Progression. Eur. Urol. 2017, 72, 835–844.
- Han, H.; Wang, Y.; Curto, J.; Gurrapu, S.; Laudato, S.; Rumandla, A.; Chakraborty, G.; Wang, X.; Chen, H.; Jiang, Y.; et al. Mesenchymal and stem-like prostate cancer linked to therapy-induced lineage plasticity and metastasis. Cell Rep. 2022, 39, 110595.
- Nouruzi, S.; Ganguli, D.; Tabrizian, N.; Kobelev, M.; Sivak, O.; Namekawa, T.; Thaper, D.; Baca, S.C.; Freedman, M.L.; Aguda, A.; et al. ASCL1 activates neuronal stem cell-like lineage programming through remodeling of the chromatin landscape in prostate cancer. Nat. Commun. 2022, 13, 2282.
- Mu, P.; Zhang, Z.; Benelli, M.; Karthaus, W.R.; Hoover, E.; Chen, C.C.; Wongvipat, J.; Ku, S.Y.; Gao, D.; Cao, Z.; et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 2017, 355, 84–88.
More
