Circular RNAs (circRNAs), a newly recognized group of noncoding RNA transcripts, have established widespread attention due to their regulatory role in cell signaling. They are covalently closed noncoding RNAs that form a loop, and are typically generated during the splicing of precursor RNAs. CircRNAs are key post-transcriptional and post-translational regulators of gene expression programs that might influence cellular response and/or function. In particular, circRNAs have been considered to function as sponges of specific miRNA, regulating cellular processes at the post-transcription stage. Accumulating evidence has shown that the aberrant expression of circRNAs could play a key role in the pathogenesis of several diseases. Notably, circRNAs, microRNAs, and several RNA-binding proteins, including the antiproliferative (APRO) family proteins, could be indispensable gene modulators, which might be strongly linked to the occurrence of diseases. In addition, circRNAs have attracted general interest for their stability, abundance in the brain, and their capability to cross the blood–brain barrier.
1. Introduction
Many studies have identified some non-coding RNAs (ncRNAs) with abnormal expressions in several diseases and/or disorders
[1]. Among them, circular RNAs (circRNAs) are a special type of endogenous ncRNAs, probably formed by back-splicing events, which have attracted more and more interest nowadays. CircRNAs are closed loop structures
[2], in which the 3′ and 5′ ends are covalently joined
[3]. The altered expression of specific circRNAs might play an important role in human diseases and/or disorders
[4]. CircRNAs were first thought to be something similar to viroids in plants
[5]. Although the abundance of a number of circRNAs is lower than their counterpart linear RNAs, circRNAs are commonly definitely expressed
[6]. In addition, circRNAs are much more stable than linear mRNAs or non-coding microRNAs (miRNAs)
[7]. The lack of a linear terminal at both the 3′ and 5′ ends might hamper their degradation by RNases, which could increase their stability in the extracellular environment, and support their helpfulness as biomarkers of diseases
[8]. In fact, it has been shown that circRNAs are more resistant than linear mRNA to the degradation of RNase R due to the lack of 5′ and 3′ ends, including the terminal 5′ caps and/or 3′ poly (A) tails
[9]. CircRNAs could play important roles in cellular processes. The first mechanism, with regards to the role of circRNAs, might be as a sponge of miRNAs
[10]. Some abundant circRNAs could associate with miRNAs through regions of complementarity, which may enable the translation of the mRNAs by taking the miRNAs away from the target mRNAs. In addition, circRNA could also bind to proteins in the related signaling pathways
[11]. Furthermore, circRNAs could be translated to protein peptides
[12]. Consequently, circRNAs might promote RNA translation and/or the stabilization of the miRNA assembly. Therefore, the specific expression of circRNAs may be different from the other classes of RNAs.
According to their splicing sequence, circRNAs can be categorized into several groups. Exon–intron circRNAs and intronic circRNAs are mainly located in the nucleus, suggesting that they may be involved in gene expression
[13]. Exonic circRNAs are the most abundant circRNAs found in the cytoplasm
[14]. A lariat structure may be formed by the covalent binding of the splice donor of the downstream exon of the precursor mRNA and the splice acceptor of the upstream exon, leading to the formation of exon–intron circRNAs and exonic circRNAs. In this model, further back-splicing by the covalent joining of the 3′ and 5′ ends might result in an intronic circRNA lariat. In some cases, exon skipping might also result in mixed circRNA lariats
[15]. Circularization might be created from the introns flanking the exons of the pre-mRNA sequence. In addition, it has been reported that the biogenesis of circRNAs could be regulated by RNA-binding proteins (RBPs)
[16]. A circular structure could be created by the connection of RBPs to introns on both flanking exons of the pre-mRNA sequence. RBPs could recognize the specific motifs of the back-splicing to form circRNAs. CircRNA may be localized in the nucleus, where it can recruit several proteins to modify the chromatin structure and/or to bind to DNA forming an RNA–DNA hybrid for the transcriptional alterations
[17]. In the cytoplasm, circRNAs have also been detected; regulating target mRNA expression by acting as miRNA sponges
[18]. CircRNAs are broadly present in tissues, blood, and urine with structural stability
[19]. Similar to the ncRNAs, circRNAs could be encumbered into exosome vesicles to facilitate cell–cell communication
[20].
Again, circRNAs have emerged as novel regulators of gene expression by sequestering miRNAs and RBPs
[21]. In addition, it has been suggested that circRNAs might play a critical role in regulating cellular events by interacting with RBPs
[22]. In cases of miRNAs, miRNAs could regulate translation and mRNA stability by binding target mRNAs in a complex with Argonaute (AGO) proteins
[23]. AGO proteins might interact with a member of the trinucleotide repeat containing six (TNRC6) family proteins to form a microRNP complex, which recruits the carbon catabolite repression 4 (CCR4)-negative on the TATA-less (NOT) complex to accelerate deadenylation with the inhibition of translation
[24]. Deletion of the poly (A)-binding protein (PABP) interacting motif (PAM2) from the TNRC6 could abolish the translational activation, suggesting the involvement of PABP in the functional process of circRNAs and/or miRNAs
[24]. Interestingly, the transducer of erbB2 1 (Tob1), a member of the antiproliferative (APRO) protein family, could simultaneously interact with the poly (A) nuclease complex CCR4-chromatin assembly factor-1 (CAF1) and the cytoplasmic PABP
[25]. In addition, the transducer of erbB2 2 (Tob2), another member of the APRO protein family, could promote deadenylation by recruiting Caf1 deadenylase onto the mRNA poly (A) tail by also interacting with PABP
[26]. The APRO family genes have been categorized in the group of immediate early growth responsive genes
[27]. The gene products might include similar molecules including pheochromocytoma cell-3(PC3)/tetradecanoyl phorbol acetate-inducible sequences 21 (TIS21)/B-cell translocation gene 2 (BTG2), B-cell translocation gene 1 (BTG1), Tob1, Tob2, abundant in neuroepithelium area (ANA)/B-cell translocation gene 3 (BTG3), PC3B and others
[27].
2. Roles of CircRNAs in Pathophysiology
In the nucleus, circRNAs can be synthesized by the reverse splicing of coding exons and/or mRNA splicing without degradation of the intron
[28][33]. CircRNAs can also regulate gene expression in the nucleus by binding to miRNA or proteins
[29][34]. Interestingly, many studies have confirmed the role of circRNA in various diseases. For example, circRNA homeodomain-interacting protein kinase 3 (HIPK3) may induce endothelial proliferation and/or vascular dysfunction in diabetic retinopathy
[30][35]. Cancer-specific circRNAs could also promote transformation and cell survival
[31][36]. In addition, circRNAs may play key roles in the progression of various diseases through their biological effects, in part, by interacting with RBPs, serving as sponges of miRNA, and/or contributing to protein coding
[32][33][34][37,38,39]. In fact, several circRNAs may be involved in regulating pathological processes
[35][40]. Furthermore, circRNAs are abundant in the brain and/or in exosome vesicles
[36][41]. Their capability to transverse the blood–brain barrier (BBB) makes them useful candidates as potential diagnostic tools for central nervous system (CNS) disorders
[36][41]. Some circRNAs’ expression increases during CNS development to raise the concentration of miRNA target sites
[37][30]. Remarkably, a large proportion of circRNAs are abundant in the brain with an unequal distribution in the neuronal compartments. CircRNAs may be involved in the regulation of circulating miRNA genes in CNS disorders such as Alzheimer’s disease
[38][42]. Furthermore, circRNAs have been recognized as potential biomarkers in other diseases including amyotrophic lateral sclerosis (ALS), diabetes, and glioblastoma
[39][43]. For example,
circSMOX RNA has been identified as a biomarker in genetic mice models of ALS with the potential for indicating disease progression
[40][44].
Interestingly, circRNAs may decrease during cell proliferation, even in some cancer cells
[41][45]. Many circRNAs with miRNA response elements have been discovered to play essential roles by acting as endogenous competitive RNAs
[10]. For example, ciRS7, a typical sponge of miR-7, contains more than 70 miR-7 binding sites
[32][37]. In addition, circSPARC may upregulate the expression of janus kinase 2 (JAK2) by competitively binding to miR-485-3p, and might augment the migration and/or invasion of colorectal cancer
[42][46]. In addition, circRNAs have been revealed to accomplish biological functions by interacting with RBPs and/or participating in protein coding
[43][47]. Furthermore, circRNAs may also regulate transcription to disturb the expression of their parental genes
[44][48]. Amazingly, the unique structure of circRNAs makes them exciting for use as potential diagnostic biomarkers even for cardiovascular diseases
[45][49]. Likewise, recent evidence has identified a crucial role of several extracellular circRNAs in alleviating damage due to cardiohypertrophy, heart failure, and myocardial infarction
[46][50]. Furthermore, several studies have reported their association with inflammatory responses; thus, influencing pathophysiological phenomena in various tissues
[47][51], which may become targets for disease therapy. CircRNA is an important cargo carried by exosomes, which could modulate gene expression by sponging certain miRNAs, regulating nuclear transcription, and competing with mRNA splicing
[48][52]. In addition, their closed loop structure determines the high biological stability of circRNAs; thus, making them a promising biomarker in clinics.
3. CircRNAs and Several Diseases
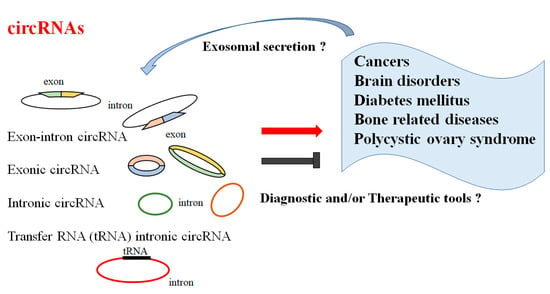
CircRNAs have attracted general interest for their stability, abundance in the brain, capability to cross the BBB, and their specific expression in several diseases. Accumulating evidence has shown that aberrant expression of circRNAs could play a key role in the pathogenesis of several diseases. Notably, circRNAs might be indispensable immune system gene modulators, which might be strongly linked to the occurrence of autoimmune disorders (Figure 1).
Figure 1. Illustration of the general functions of circular RNAs (circRNAs). Functions of circRNAs have been proposed in several diseases including cancers, brain or CNS disorders, several types of diabetes mellitus, bone-related diseases, and polycystic ovary syndrome. Consequently, certain circRNAs could be diagnostic and/or therapeutic tools for these diseases. The arrowhead means stimulation and/or augmentation, whereas the hammerhead represents inhibition.
4. Mechanism of circRNAs’ Action with PABP and APROs
As shown here, a lot of evidence suggests that circRNAs may play a key role in disease initiation and/or progression. In addition, circRNAs may play a key role in the proliferation, differentiation, and/or apoptosis of various cells in those diseases. Given the diverse roles attributed to circRNAs and miRNAs, the molecular biological mechanisms of these RNAs might be quite elaborate. Again, circRNA could regulate gene transcription, alternative splicing, molecular sponges of miRNA, RNA-binding proteins, and/or protein translation. In translation, miRNAs might be bound with AGO proteins in the 3′ UTR of complementary mRNA sites. The AGO-miRNA activity might be further modulated by adjacent RBPs by interacting with target proteins. A single circRNA can bind to one or more miRNAs through its circular sequences. Accordingly, circRNAs could also interact with transcription machinery including RNA Pol II and/or U1 snRNP to promote their parent gene expression in the nuclei. Probably, long non-coding RNA could act as a competitive endogenous RNA to compete for miRNA binding. Some p-element induced wimpy testis (piwi)-interacting RNAs could also interact with BTG1 expression
[49][120]. Piwi proteins have been shown to be a subfamily of Argonaute proteins that maintain germ cells in eukaryotes. The knockdown of piwil1, a member of the piwi-like protein family, could increase the expression of the transcriptional co-regulator BTG2
[50][121]. Piwi-interacting RNAs and PIWI proteins may be essential in cells to repress transposons and/or to regulate mRNAs. Consequently, circRNAs could regulate mRNA stability and immune cell death by binding to specific RBPs. In addition, circRNAs could indirectly regulate gene expression by binding targeted miRNAs in cells including immune cells
[51][122]. Certain circRNAs might also block the translation of the host gene by binding to the adjacent PABP
[52][123]. The association of circRNAs and PABP might affect the combination of PABP and eIF4G on the 5′ cap region of mRNA, which specifically affects the translation and/or the expression of certain mRNA
[52][123] (
Figure 2).
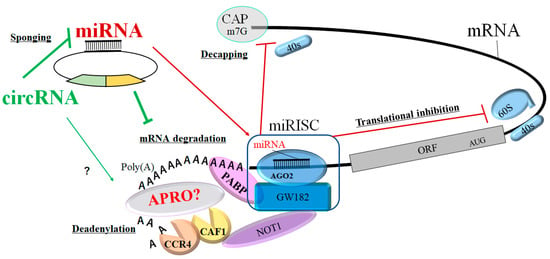
Figure 2. Schematic representation of circRNA- and/or miRNA-mediated functional inhibition of mRNA translation. The AGO2 protein interacts with GW182 constructing the miRNA-loaded RNA-induced silencing complex (miRISC), which may facilitate the deadenylation and/or mRNA degradation process by CAF1/CCR4/NOT1 with the PABP and APRO protein complex. Consequently, the circRNA and/or miRNA could play an active role in regulating post-transcriptional gene expression via the decapping, translational inhibition, deadenylation, and degradation of mRNA. The CAF1/CCR4/NOT1 complex is recruited to the 3′ UTR of specific mRNAs through an interaction with the PABP protein. APRO family proteins might also interact with PABP to recruit the CAF1/CCR4/NOT1 complex. The arrowhead means stimulation, and the hammerhead represents inhibition. Note that some critical pathways have been omitted for clarity. Abbreviations: ORF, open reading frame; miRISC, microRNA-induced silencing complex, “?” means for
the our
esearchers' speculation.
Some circRNAs are subject to endoribonucleolytic cleavage with the target of mRNA and/or miRNA
[53][124]. Several members of the APRO family are also shown to be implicated in cytoplasmic mRNA deadenylation and its turnover
[54][125]. The N-terminal conserved APRO domain is capable of binding to DNA-binding transcription factors as well as the deadenylase subunits of the CCR4/NOT complex
[55][126]. Likewise, some of the APRO family proteins could interact with the poly(A) nuclease complex CCR4/CAF1 and the cytoplasmic PABP
[25][56][25,127]. In addition, it has been shown that the Tob1 and Tob2 proteins contain an extra-long C-terminal domain with two PAM2 motifs
[57][128]. These APRO proteins (Tob1 and Tob2) could interact with CAF1 and PABP simultaneously, which might stimulate the deadenylation of mRNA
[56][127]. Interestingly, the antiproliferative effects of Tob1 have been suggested to be involved in the exploitation of the CAF1/CCR4 deadenylase complex
[58][129], suggesting that APRO proteins could exert their antiproliferative activity by modulating the turnover of mRNA
[59][130]. In fact, BTG2 could also interact with CAF1 deadenylase through its APRO domain to control cell proliferation
[60][131]. It has been shown that mRNA destabilization by BTG1 and/or BTG2 may sustain cell quiescence
[61][132]. Hence, circRNAs and miRNAs could inhibit mRNA expression by base-pairing to the 3′ UTR of the target mRNAs, which consequently inhibits translation by initiating poly(A) tail deadenylation and mRNA destabilization with APRO family proteins
[62][133]. In fact, the miRISC could interact with PABP, CAF1, and CCR4 deadenylases
[63][134]. Importantly, a core component of the miRISC could interact with the PABP and APRO family proteins, which may be compulsory for the miRNA-mediated deadenylation
[63][134]. Since the APRO family proteins have the potential to interact with the CCR4/CAF1 complex, APRO family proteins could be a key modulator of circRNAs- and/or miRNAs-function. Therefore, APRO and the CCR4/CAF1 protein complex might be a multifunctional regulator that plays an important role in multiple cellular processes in eukaryotes
[64][135]. The expression of APRO family proteins may be also regulated by certain circRNAs and/or miRNAs
[49][65][120,136].