Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Wiwin Effendi and Version 3 by Sirius Huang.
Epigenetics describes molecular missing link pathways that could bridge the gap between the genetic background and environmental risk factors that contribute to the pathogenesis of pulmonary fibrosis. Specific epigenetic patterns, especially DNA methylation, histone modifications, long non-coding, and microRNA (miRNAs), affect the endophenotypes underlying the development of idiopathic pulmonary fibrosis (IPF).
- epigenetics
- epigenomics
- DNA methylation
- long noncoding
- microRNA
- histone modification
- idiopathic pulmonary fibrosis
- chronic pulmonary diseases
1. Introduction
Idiopathic pulmonary fibrosis (IPF) is a chronic, devastating, and irreversible lung disease that is characterized by microinjury-induced alveolar epithelial cell stress, progressive pathogenic myofibroblast differentiation, imbalanced macrophage polarization, and the extensive deposition of the extracellular matrix (ECM) [1][2][3][1,2,3]. The progression of patients with IPF is associated with lung function decline, progressive respiratory failure, high mortality, recurrent acute exacerbations, and an overall poor prognosis [4][5][6][4,5,6]. The morphological hallmark of IPF on histopathological and/or radiological is usual interstitial pneumonia (UIP), composed of heterogeneous areas of normal-appearing lung intermixed with collagenized fibrosis in sub-pleural and paraseptal, a honeycombing pattern, and ECM-producing myofibroblasts termed fibroblast foci (FF) [7][8][7,8].
A recent hypothesis stated that recurrent injuries drive the aberrant activation of epithelial cells to transdifferentiate into fibroblast epithelial-mesenchymal transition (EMT), which might induce fibrosis independently of inflammatory events [9][10][9,10]. Even though there is no implicit mechanism, several shreds of evidence emphasize that alveolar epithelial injury induced by environmental triggers results in lung fibrosis. Recurrent microenvironment injury on senescent epithelial cells in genetically susceptible individuals leads to the aberrant activation of fibroblasts, accumulating ECM, and fibrosis [11].
The pathogenic mechanisms involved in the initiation, development, and progression of IPF are unclear. However, many studies have demonstrated that dynamic interactions of genetic susceptibility, environmental factors, and host risk factors in older individuals contribute to epigenetic pro-fibrotic reprogramming, resulting in the development of IPF [12]. Hey et al. found a strong association between the microenvironment-driven epigenetic changes that could induce macrophage inflammation and polarization [13].
Omics-based approaches, including high-throughput technologies that provide snapshots of a holistic view of the molecules that make up a cell, tissue, or organism, consist of (1) genomics, measuring deoxyribonucleic acid (DNA) sequence variation; (2) epigenomics, focusing on the genome-wide characterization of reversible modifications of DNA or DNA alterations; (3) transcriptomics, evaluating the standard of ribonucleic acid (RNA) expression; (4) proteomics, determining protein expression or its chemical changes; (5) metabolomics, assessing metabolite/small molecule levels; and (6) microbiomics, investigating all the microorganisms of a given community [14][15][14,15]. Pulmonary disease omics studies mainly focus on tissue- and cell-specific omics data and have identified several fundamental mechanisms that underlie pulmonary biological processes, disease endotypes, and appropriate novel therapeutics for selected individuals [16].
Epigenomics is a technique for analyzing gene expression through epigenetic mechanisms, including DNA methylation, RNA, and histone modification [17]. These components interact and stabilize each other; therefore, the disruption of epigenetic nucleosomes can lead to their inappropriate expression, resulting in epigenetic disorders [18]. Epigenetics and epigenomics help explain how our environment affects our phenotype. Epigenomics studies commonly use methods such as Hi-C, a comprehensive technique developed to capture chromosome conformation, and another tool for whole genome methylation profiling: MBD-isolated genome sequencing (MiGS) [19]. Essential technical and experimental parameters that should be considered when designing epigenomic experiments are reviewed in detail by these authors [20].
2. DNA Methylation in Fibrosis
Extensive alterations in DNA methylation profiles are known to be involved in the pathogenesis of pulmonary fibrosis. DNA methylation microarray demonstrated a higher DNA methyltransferase expression in lung tissue samples of IPF patients [21][81]. IPF fibroblasts exhibited significant heterogeneity in global DNA methylation patterns compared to non-fibrotic control cells [22][82]. Additionally, a study identified extensive alterations in gene expression-associated DNA methylation that were involved in fibroproliferation [23][83]. Recently, a genome-wide DNA methylation study found that CpGs methylation was responsible for cell adhesion, molecule binding, chemical homeostasis, surfactant homeostasis, and receptor binding categories in the pathogenesis of IPF [24][84]. Global methylation patterns in IPF are very different from that of the control samples and significantly overlap with methylation changes observed in lung adenocarcinoma samples [25][85]. DNA methylation biomarkers also overlap between IPF, other ILD, cancer, and COPD; hence, it is unlikely that molecular discrimination between these diseases can be achieved using a single marker [26][86]. Interestingly, low-methylation lung squamous cell carcinoma (SCC) significantly correlated with IPF to show a bad outcome [27][87]. Cigarette smoking-associated aberrant methylation contributed to pulmonary fibrosis [28][88]. Many cell types are well known to be involved in the process of pulmonary fibrosis. The essential cells driving the development and progression of pulmonary fibrosis are epithelial, fibroblasts and myofibroblasts, and alveolar macrophages. Epigenetic regulation has a variety of ways. This discussion focuses on DNA methylation regulation in fibroblasts, epithelial, and macrophages.2.1. DNA Methylation and EMT
EMT is a cellular and molecular process in which epithelial cells lose their epithelial identity (apical-basal polarity and adhesion), which is characterized by down-regulated epithelial markers, including E-cadherin, occludin, and claudin-1. In contrast, fibroblast-specific genes, such as α-SMA, N-cadherin, fibroblast-specific protein 1 (FSP-1), and type I collagen, are up-regulated [10][29][10,89]. A hallmark of EMT is the repression of E-cadherin: a transmembrane glycoprotein encoded by the epithelial marker gene CDH1. DNA methylation might be involved in regulating EMT. During EMT induction, EMT-transcriptional factors (EMT-TFs), including ZEB1, SNAIL, and TWIST, interact with DNMTs and undergo the hypermethylation of CpG islands in the CDH1 promoter [30][31][90,91]. The DNA hypermethylation of the CDH1 through DNMTs was correlated with tumor progression [30][32][90,92]. In addition, the knockdown of DNMTs was equated with the transfection of siDNMT1, DNMT3a, or DNMT3b, which reversed the TGF-β1-induced suppression of E-cadherin expression and the induction of α-SMA, vimentin, and fibronectin expression [33][93]. In normal lung epithelial wound healing, this process ends with the apoptosis of myofibroblasts and inflammation reduction. Yet, aberrant responses to tissue injury may turn into lung fibrosis. Bidirectional EMT cross-talk assists the pro-fibrogenic positive feedback loop, while epithelial cells become “vulnerable and sensitive to apoptosis” and myofibroblasts become “apoptosis-resistant and immortal”, resulting in fibrosis progression instead of wound resolution [34][94]. DNA methylation mediated the down-regulation of genes involved in apoptosis. The hypermethylation of the Caspase 8 promoter by DNMT1 and DNMT3b was associated with apoptosis resistance in cancer cells [35][36][95,96]. Cisneros et al. showed the silencing expression of pro-apoptotic p14ARF in the fibroblast of the patient with IPF due to hypermethylation [37][97]. Along with these changes, DNA methylation was linked with the dysregulation of apoptosis and EMT, leading to the development of lung fibrosis processes (Figure 12).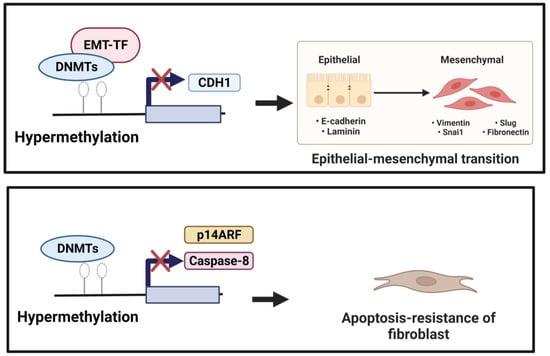
Figure 12. (1) Induction of EMT. Down-regulation of CDH1 (anti-fibrotic factor) is mediated by EMT-TF, which recruits DNMTs to the CDH1 promoter performing focal hypermethylation of the CpG islands in the CDH1 promoter, leading to decreased gene expression of CDH1. (2) Reduction in apoptotic activity. Hypermethylation mediated by DNMTs of the pro-apoptotic factors, including Caspase 8 and p14 ARF, could underlie resistance to fibroblast apoptosis.
2.2. DNA Methylation and Myofibroblast Differentiation
Under normal physiology, in response to epithelial injury, the cell releases various pro-inflammation and fibrotic cytokines that are responsible for local inflammation and the activation of fibroblasts. Fibroblasts are mesenchymal cells that are recruited to and accumulate at the injured site and undergo a change in phenotype to highly proliferative and contractile myofibroblasts. Lung myofibroblasts are heterogeneous in terms of their origins. The predominant sources of myofibroblasts are resident fibroblasts (lipofibroblasts, matrix fibroblasts, and alveolar niche cells) and pericytes (residing within basal membranes or perivascular linings), along with minor sources such as hematopoietic CXCR4+ fibrocytes, alveolar epithelial cells (AECs), endothelial cells (ECs), and mesenchymal stem cells (MSCs) [38][98]. Under the influence of environment-specific factors, cytokines, such as TGF-β, fibroblast-to-myofibroblast transition/transdifferentiation (FMT) changes lung resident fibroblast behavior/phenotypes to another type of normal differentiated cell and the downstream effects at the tissue level [39][99]. Myofibroblasts, characterized by the expression of contractile α-SMA, regulate connective tissue remodeling by producing and modifying ECM components, such as fibronectin and collagen [40][100]. Furthermore, one of the mechanisms that may account for FMT is DNA methylation. DNA methylation modulates myofibroblast differentiation (Figure 23). Several genes and signaling pathways, such as those involving ErbB, focal adhesion, and MAPK, underlie mechanisms for DNA methylation alteration in myofibroblast differentiation and influence the synthesis of ECM [41][101]. DNA methylation regulates gene expression to facilitate the formation of fibroblastic foci and lung fibrosis [42][47]. DNMT is an essential regulator of α-SMA gene expression during myofibroblast differentiation. The specific effect of DNA methylation in regulating the α-SMA gene is unknown. During the development of cardiac fibrosis, He et al. found that TGF-β could inhibit DNMT1-mediated DNA methylation expression, leading to the overexpression of α-SMA [43][102]. TGF-β1 also regulates lung fibroblast differentiation through the hypermethylation of the “fibrosis suppressor” gene, Thymocyte differentiation antigen-1 (THY-1); therefore, DNMT1-attenuated TGF1-mediated THY-1-inducing α-SMA fiber formation is silenced [44][103]. In addition, the expression of α-SMA was significantly increased by adding IL-6 [45][104]. A recent study revealed that DNMTl knockdown suppressed the DNA methylation-related myofibroblast phenotype via inhibiting an IL-6/STAT3/NF-κβ positive feedback loop [46][105].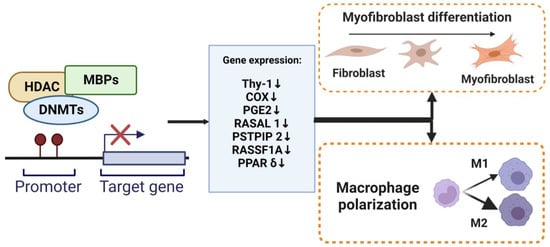
Figure 23. DNA methylation regulate myofibroblasts and macrophages. The methylation of promoter DNA can repress gene expression by MBPs, recruiting specific macromolecular complexes that contain histone deacetylases (HDAC), DNMTs, and other transcriptional co-repressors (Co-Rep). Several genes that function as an anti-fibrotic and are associated with myofibroblast differentiation and macrophage M2 polarization have been shown to undergo silencing due to DNA hypermethylation.
