You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 1 by Paul David and Version 2 by Peter Tang.
Gastrointestinal (GI) cancers are a common cancer, affecting both men and women, normally diagnosed through tissue biopsies in combination with imaging techniques and standardized biomarkers leading to patient selection for local or systemic therapies. Liquid biopsies (LBs)—due to their non-invasive nature as well as low risk—are the current focus of cancer research and could be a promising tool for early cancer detection and treatment surveillance, thus leading to better patient outcomes.
- liquid biopsy
- circulating tumor cells (CTCs)
- circulating tumor DNA (ctDNA)
- tumor exosomes
- tumor-educated blood platelets (TEPs)
- organoids
- gastrointestinal cancer
1. Introduction
Gastrointestinal (GI) cancers are responsible for more cancer-related deaths than lung and breast cancer. Colorectal cancer (CRC) is the major type of GI cancer, with 1.9 million new cases diagnosed worldwide in 2020, making it after lung and breast cancer the third most common cancer of all organs. According to the International Agency for Research on Cancer, in the same year, 1.1 million new cases of gastric cancer, 900,000 new cases of liver cancer, 600,000 new cases of esophageal cancer, and 500,000 new cases of pancreatic cancer were diagnosed across the globe [1].
Although the prognosis of many GI cancers has improved over the past decades [2][3][2,3], a late cancer diagnosis is still the leading reason for cancer-related deaths among all GI cancers [4]. Current research focuses therefore on improving early cancer diagnosis, possibly leading to better outcomes among all GI cancers [5][6][5,6]. So far, endoscopic or CT-guided solid biopsies in combination with so-called serum-based tumor biomarkers are primary methods for the diagnosis of GI cancers [7]. Thereby, solid biopsies are considered the gold standard strategy capable of classifying tumors, identifying the mutational status, and providing prognostic information. However, these methods have some limitations, e.g., obtaining insufficient or inaccurate tissue samples possibly leading to false-positive or false-negative results. In addition, tissue biopsies might cause harm to the patient. However, recent studies suggest tissue biopsies taken from a single cancer nodule or single metastatic lesion may fail to represent the entire tumor heterogeneity within the patient, possibly being one of the main reasons for the failure of current targeted therapies [8][9][10][11][12][13][8,9,10,11,12,13]. To date, several serum-based biomarkers such as carcinoembryonic antigen (CEA), carbohydrate antigen 19-9 (CA19-9), carbohydrate antigen 72-4 (CA72-4), carbohydrate antigen 125 (CA125), and alpha-feto protein (AFP) have been identified and widely used for diagnosis, prognosis, and monitoring of potential recurrence of GI cancers [14][15][14,15]. Although, due to the limit of specificity and sensitivity most of these biomarkers are not useful for early cancer detection [16]. Therefore, LB emerged as a promising tool for early detection, treatment selection, and real-time prognosis.
In contrast to solid biopsy, LB is a minimally invasive approach enabling the real-time monitoring and early uncovering of alterations in cells or cell products shed from malignant lesions into the body fluids (Figure 1). LB analysis can identify multiple heterogeneous resistance mechanisms in single patients compared to solid biopsy. Furthermore, LB facilitates the choice of the right treatment and observation of the treatment response. Due to the minimally invasive nature of LB, the resulting complications from obtaining solid biopsies could be prevented. A typical LB sample is taken from any biological fluid such as blood, saliva, cerebrospinal fluid, or urine. LB materials derived from peripheral blood have been investigated extensively. LB analysis from blood contains enrichment and isolation of CTCs, circulating blood platelets, ctDNA, and other tumor genetic material such as extracellular vesicles. As of today, several LB technologies have been approved by the United States Food and Drug Administration (FDA) for malignancies such as metastatic lung, breast, prostate, or colorectal cancer: CELLSEARCH CTC test using circulating tumor cells from Veridex, Guardant360 CDx, and FoundationOne Liquid CDx using circulating cell-free DNA (cfDNA) and next-generation sequencing to detect tumor-specific mutations.
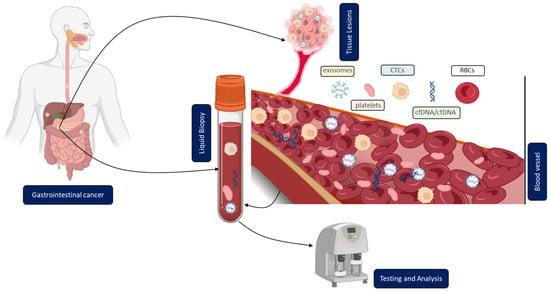
Figure 1. Clinical application of liquid biopsy (LB) in gastrointestinal cancer (GICs). Circulating tumor cells (CTCs), cell-free or circulating tumor DNA (cfDNA/ctDNA), tumor-educated platelets (TEP), exosomes, and RBCs in the blood of GICs patients can be used as potential biomarkers for LBs and their expression levels can be measured to determine the clinical status of GICs patients.
2. Overview of Different Methodologies and Their Current Clinical Application
2.1. Circulating Tumor Cells (CTCs)
CTCs are tumor cells, shed from a primary tumor. They can enter the bloodstream or lymphatic system, potentially spreading into distant organs possibly leading to metastases [17][18][17,18]. Nevertheless, only a minority of CTCs become solid metastatic lesions because of a complex sequence of events needed, i.e., the detachment from the primary tumor, migration through the circulating blood, immune escape, and survival. It remains unclear how the detachment process from the primary tumor tissue takes place. Evidence supports the involvement of epithelial to mesenchymal transition, by which transformed epithelial cells can acquire the ability to invade, resist apoptosis, and disseminate. This could be the main driver for the detachment of tumor cells from the primary tumor [19][20][21][22][23][24][19,20,21,22,23,24]. Other reports hypothesize that cells split into different clusters [25]. It is noteworthy that gastrointestinal cancers compared to breast cancer have lower numbers of CTCs in peripheral blood due to portal vein circulations and a steady ‘first-pass effect’ in the liver [26]. Therefore, portal vein blood might be a unique sample site to isolate CTCs from gastrointestinal cancers. It has already been shown that the number of enriched CTCs from portal vein blood is higher than in the systemic circulation [27][28][27,28]. Portal vein blood can be collected intraoperatively or even by endoscopic ultrasound (EUS)-guided sampling [29].
In addition to the number of CTCs, the analysis of physical (size, density, and electric charge) and biological (cell surface expression) properties could play a crucial role in future clinical use [30].
