RNF11 (Ring Finger Protein 11) is a 154 amino-acid long protein that contains a RING-H2 domain, whose sequence has remained substantially unchanged throughout vertebrate evolution. RNF11 has drawn attention as a modulator of protein degradation by HECT E3 ligases.
- Ring Finger Protein 11
- HECT ligases
- ubiquitination
Note:Dear author, the following contents are excerpts from your papers. They are editable. And the entry will be online only after authors edit and submit it.
1. Introduction
The covalent attachment of ubiquitin to a variety of cellular proteins is a common post-translational modification (PTM) in eukaryotic cells. In addition to providing a signal for proteasomal degradation, ubiquitination is associated with different outcomes, from changing protein localization to modulating the biochemical properties of the target protein [1]. The conjugation process leads to the formation of an isopeptide bond between the C-terminus of ubiquitin and ε-amino group of a lysine residue [2]. Ubiquitin conjugation involves the sequential transfer of ubiquitin through an enzymatic cascade that consists of a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3). E3 largely determines the high degree of specificity and selectivity for the target proteins and shows the greatest diversity among the ubiquitination machinery. RING (really interesting new gene) ligases, which represent the largest family of E3-ligases that consist of over 600 genes in human, are commonly considered as both scaffolds and allosteric activators of the conjugating E2 enzymes. Indeed, their RING domain brings in close proximity the specific E2 to the substrate and then promotes the direct transfer of ubiquitin from the E2 to the target protein [3]. Two other groups, called HECT (Homologues to the E6AP carboxyl terminus) and RBR (RING between RING) ligases, make up roughly 5% of the total number of known E3-ligases. The conjugation of ubiquitin by HECT and RBR E3-ligases takes place in two steps: ubiquitin is first transferred from the E2 to an active-site cysteine residue on the catalytic domain of the ligase and then the target substrate [4][5][6][4,5,6]. Despite their limited number, their well-known involvement in several human disease-related processes has called for a deeper understanding of their working mechanisms.
2. RNF11, A Multifunctional RING-H2 Ligase
2.1. Domain Organization and Cellular Localization of RNF11
RNF11 is a 154-amino acids protein that contains a canonical RING-H2 finger motif (C3H2C2, 99–140aa) at the carboxyl-terminal end (Figure 1). The RING-H2 domain appears to be a stable folded monomeric domain showing structural similarities with the RING1 domain of RBR E3s [7][12]. The full-length protein contains disordered regions, which mainly involving the N-terminal end and the short C-terminal tail following the RING domain (aa 141–154) [7][12]. However, the structure of both terminal ends is predicted to be highly influenced by the presence of post-translational modifications and the interaction with binding partners (see below).
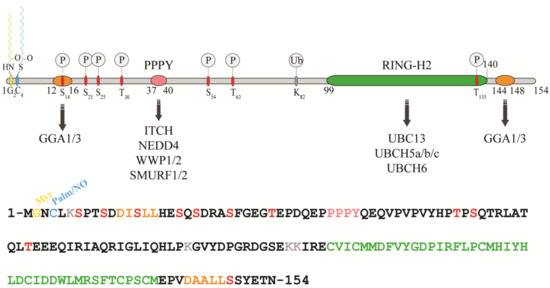
Figure 1. Protein sequence, domain organization and binding partners of the RING-H2 E3-ligase RNF11. Residues identified as target sites for post-translational modifications are shown. Colours legend: the PPY motif is coloured in pink, the RING-H2 domain in green, the myristoylation site is in yellow, the palmitoylation/nitrosylation site in light blue, the di-leucine motifs are in orange, the phosphorylation sites are in red, and the lysine residues are in grey.
The remarkable aspect of RNF11 is the presence of a PY (37-PPPY-40) motif upstream to the RING-H2 domain that allows for RNF11 to interact with the WW-domains containing proteins of the NEDD4 family of HECT-E3 ligases (among them: ITCH, NEDD4, SMURF1/2, and WWP1/2) [8][9][10][11][12][13][13,14,15,16,17,18]. This feature makes RNF11 the only RING ligase potentially capable of interacting with all the members of the NEDD4-like family. In addition, a putative ubiquitin-interacting motif (UIM) (corresponding to residues 67–74), located between the PY and the RING-H2 motif, has been predicted [14][19]. Nevertheless, the presence of a glycine (Gly) instead of the highly conserved serine (Ser) makes the effective functionality of this motif controversial. Accordingly, recent data show that the UIM of RNF11 does not bind ubiquitin in vitro, which leaves open the question whether it acts as a functional ubiquitin binding site in vivo [7][12]. Because the non-covalent interaction between E3 enzymes and ubiquitin has clearly been shown to influence the enzymatic activity of almost all the main classes of ligases [15][16][17][18][19][20][21] [20,21,22,23,24,25,26], clearly defining the ability of RNF11 to bind ubiquitin would provide useful hints regarding its regulatory mechanism.
In growing cells, RNF11 shows an intracellular vesicular distribution, which, in a steady state, mainly corresponds to the early and recycling endosomes. The access to these compartments appears to be strictly regulated by multiple sorting determinants. The N-terminal end of RNF11 undergoes a double acylation during protein maturation [12][17]. Immediately after the removal of the first methionine, which takes place during translation, a myristoyl group is covalently and irreversibly attached to Gly2 by an N-myristoyl transferase, followed by palmitoylation of the proximal cysteine 4 (Cys4), a reversible modification that occurs in the Golgi compartment [22][23][27,28]. The double N-terminal acylation is a pre-requisite for membrane localization [12][17]. Additional signals and protein-protein interactions allow for the intracellular trafficking of RNF11 and its access to the early and recycling endosomes [12][17]. More specifically, two acidic di-leucine motifs are the sorting determinants that drive the intracellular traffic of RNF11. The N-terminal motif (15-LL-16) is preceded by two aspartic residues, Asp11 and Asp12. These residues give rise to two partially overlapping acidic di-leucine sorting motifs: the DxxxLL consensus sequence (residues 11–16) mediates the interaction with the clathrin adaptor protein 2 (AP-2) complex, while the DxxLL motif (residues 12–16) interacts with the VHS (Vps27p, Hrs, and STAM) domains of the Golgi-localized, gamma adaptin ear-containing, ARF-binding (GGA) protein family. The di-leucine couple acts as a sorting signal at both the trans-Golgi Network (TGN) and at the plasma membrane. Conversely, a C-terminal acidic di-leucine motif (144-DAALL-148) seems to rather regulate the distribution of RNF11 inside the endosome compartment, between the early endosome and the recycling routes [13][18].
2.2. RNF11 in Intracellular Traffic, Signal Transduction and Oncogenesis
Within higher eukaryotic cell membranes, traffic pathways connect various membrane organelles and maintain cellular homeostasis by retaining the correct complement of proteins and lipids. The exocytic/secretory pathway carries newly synthetized proteins and lipids from the endoplasmic reticulum (ER) through the Golgi to the plasma membrane. The endocytic pathway internalizes proteins from the extracellular environment or from the plasma membrane to the early endosomes. Here, cargos are either sorted to the recycling endosome, to be brought back to the plasma membrane, or they are delivered to the late endosomes, and finally to the lysosomes for degradation. Membrane-bound vesicles mediate these trafficking pathways, and sophisticated machineries, as well as complex mechanisms of regulation are required in order to ensure efficiency and specificity in the selection and delivery of cargos.
Several lines of evidence strongly support the notion that RNF11 does not act simply as a cargo of membrane adaptors, but it actively participates in controlling membrane traffic processes. Notably, RNF11 gene silencing results in a strong inhibition of ER-to-plasma membrane transfer of the well-characterized secretory cargo membrane protein tsO45G, without affecting the morphology of the ER and Golgi complex [24][56]. The molecular mechanism behind this evidence is still unknown; however, it has been recently reported that, under continuous Epidermal Growth Factor (EGF) stimulation, the overexpression of RNF11 up-regulates the mRNA levels of SEC23B, SEC24B, and SEC24D, which are components of the ER export machinery specifically required for the ER export of newly synthetized EGFR(Epidermal Growth Factor Receptor). On the contrary, RNF11 knockdown down-regulates them [25][32]. Understanding how RNF11 regulates the levels of these proteins needs further investigation. However, there might be a possible direct role of this E3-ligase in gene transcription. Indeed, RNF11 has been shown to bind DNA [10][15], and prolonged EGF treatment has resulted in the nuclear translocation of RNF11 [25][32].
Other data suggest a regulatory role for RNF11 in endocytic trafficking. The overexpression at middle and high levels of wild-type RNF11 causes the early endosome vesicles to swell. Moreover, cells overexpressing the RING domain inactive mutant of RNF11 lose the integrity of early and late endosomes and show a severe impairment of transferrin uptake [18]. Finally, Kostaras and colleagues demonstrated that RNF11 silencing decreased the degradation of activated EGFR and increased EGF recycling [26][27][30,57]. Similar effects have been described as a consequence of the silencing of well-known components of the trafficking machinery [27][28][29][57,58,59], which suggests that RNF11 could also be part of it.
The endocytic route is essential for the signal termination of membrane receptors and is strictly regulated by ubiquitination at different levels. Once activated by their cognate ligands, receptors are rapidly ubiquitinated. This represents a crucial signal, as it sorts receptors to lysosomes for degradation via the Endosomal Sorting Complex Required for Transport (ESCRT) pathway [30][60]. This pathway comprises four multi-subunit protein complexes (ESCRT-0, -I, -II, -III), which act in concert to direct ubiquitinated receptors along the degradative route [31][61]. It has been proposed that RNF11 participates structurally and functionally to the ESCRT-0 complexes. The E3-ligase interacts with the ESCRT-0 subunits HRS (Hepatocyte growth factor-regulated tyrosine kinase substrate), EPS15 (epidermal growth-factor receptor substrate protein 15), and STAM1/2 (signal transducing adaptor molecule 1/2) [9][14]. Moreover, RNF11 interacts with the SMAD anchor for receptor activation (SARA) protein, which possibly mimics the function of HRS by assembling an alternative ESCRT-0 complex [32][62]. In this context, RNF11 may have a role as a fine tuner of ubiquitination. Indeed, the sorting activity of ESCRT complexes is strictly controlled by the ubiquitination of their subunits. Many of these proteins are subject to mono- or poly-ubiquitination that is predicted to induce an autoinhibitory conformation preventing the ESCRT from recognizing ubiquitinated cargos. Thus, the regulation of the ubiquitination status of the sorting machinery is crucial for the efficient sorting of activated receptors to the lysosome and for the consequent signal termination.
2.2.1. The TGF-Beta Signaling Pathway
The signaling pathway of the transforming growth factor beta (TGF-β) is one of the major regulators of cell communication in the adult organism as well as in the developing embryo. It is involved in the regulation of crucial cellular processes, such as cell growth, differentiation, apoptosis, and cellular motility. Alterations in TGF-β signaling have been implicated in many diseases, particularly cancer, where it plays a dual role as tumor suppressor in early-stage disease and tumor promoter in advanced malignant disorders [33][64].
RNF11 emerges as a positive regulator of TGF-β signaling (Figure 2).
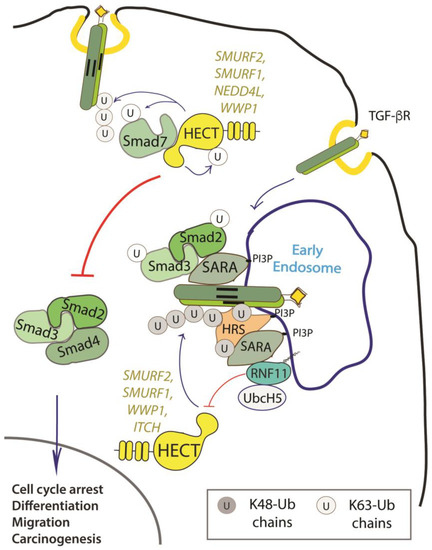
Figure 2. Role of RNF11 in the transforming growth factor beta (TGF-β) signaling pathway. Following ligand binding, the ubiquitination of the TGF-b receptor promoted by the SMURF2-SMAD7 E3 ligase complex targets the receptor to the lysosome or the proteasome for degradation. From the early endosomes, the TGF-b receptor can be sorted to recycling endosomes and return back to the cell surface or it can be driven to late endosomes and lysosomes for degradation. The activated receptor complex recruits and phosphorylates the receptor-regulated SMAD (R-SMADs, e.g. SMAD2 and SMAD3). Association of TGF-b receptors with early endosomes adaptor proteins such as SMAD anchor for receptor activation (SARA), leads to the enhancement of TGF-ß -induced SMAD activation and the subsequent propagation of SMAD-dependent signaling. Activated R-SMADs dissociate from the TGF-β receptors and associate with Smad4, a common-partner for all R-Smads (Co-SMAD). These complexes translocate into the nucleus where they regulate transcription of TGF-b family target genes by binding transcriptional factors, transcriptional coactivators and corepressor. Posttranslational modifications of SMAD proteins, among which ubiquitination by NEDD4 family of E3-ligases, provide an additional level of regulation of the TGF-β signaling, controlling protein stability and function.
2.2.2. The EGF Receptor Signaling Pathway
The EGF receptor is a membrane tyrosine kinase that initiates cell growth and proliferation. Once activated by ligand binding, EGFR is rapidly internalized and routed to lysosomes for degradation. As previously described, receptor ubiquitination is a crucial signal that commits EGFR to lysosomal degradation and allows for signal termination. The deregulation of EGFR signaling is indeed a significant feature in different stages of oncogenesis and in several types of cancer [34][68]. Evidence accumulated so far implicated RNF11 in the downregulation of EGFR signaling by promoting lysosomal degradation (Figure 3).
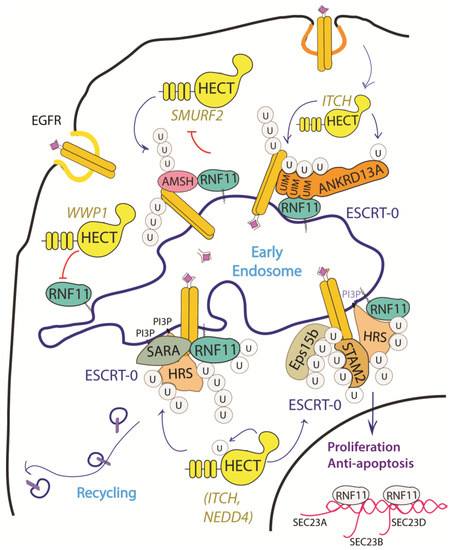
Figure 3. Graphical representation of the role of RNF11 in the EGFR signaling pathways. The Epidermal Growth Factor (EGF) receptor is a membrane tyrosine kinase that initiates cell growth and proliferation. Following ligand binding, the EGF receptor undergoes endocytosis via clathrin-dependent and clathrin-independent pathways. Following ligand stimulation, the EGF receptor undergoes endocytosis via clathrin-dependent and clathrin-independent pathways. Both routes require essential ESCRT-0 components, such as EPS15 and Epsin, which recognize the ubiquitin moieties conjugated on the receptor cytoplasmic tails following ligand stimulation. The ANKRD13 family of proteins could act as alternative ESCRT components that drive the receptor towards the endocytic routes.
The depletion and overexpression of RNF11 have both been shown to impair ligand-induced EGFR degradation and to increase EGFR recycling [25][32]. This suggests that RNF11 exerts its function in the early phases of receptor endocytosis, during which endosomal sorting occurs and EGFR is committed for degradation or recycling. Moreover, RNF11 overexpression causes a reduction of EGFR levels and receptor hyper-ubiquitination following EGF induction [35][31].
Several mechanisms have been investigated in order to clarify the role of RNF11 in the EGFR pathways, however a comprehensive framework that integrates all of them is still missing (Figure 3). RNF11 binds the DUB protein AMSH and promotes its degradation by the E3-ligase SMURF2-mediated ubiquitination [10][15]. It has been shown that AMSH interacts with the ESCRT-0 STAM-1 subunit and favours recycling of activated EGFR by promoting receptor deubiquitination. Coherently, AMSH knockdown enhances EGFR degradation [36][69]. Therefore, RNF11-mediated AMSH degradation might increase EGFR ubiquitination, clearly committing activated receptor to lysosomes.
Different HECT ligases have been demonstrated to inhibit EGFR degradation and promote cell proliferation. In particular, WWP1 upregulates EGFR levels in a ligase activity independent manner [37][39]. RNF11 associates with WWP1 and the two ligases reciprocally ubiquitinate each other in cells without affecting their degradation. The transient overexpression of WWP1 promotes cell proliferation, while WWP1 knockdown significantly suppresses cancer cell proliferation and induces apoptosis [38][31][40,61]. Interestingly, the depletion of RNF11 neutralizes the decrease of EGFR levels that are induced by WWP1 knockdown, which suggests that WWP1 could promote cell proliferation by inhibiting RNF11 [37][39][39,70]. Conversely, NEDD4 and ITCH HECT-E3-ligases have been demonstrated to inhibit EGFR degradation by ubiquitinating multiple adaptor proteins essential for the endocytosis of ligand-activated EGFR [13][32][40][41][42][43][18,62,71,72,73,74]. Again, RNF11 might counteract this by inhibiting HECT-E3 ligases activity.
2.2.3. The NF-kB Signaling Pathway
A third pathway that involves RNF11 is the Nuclear factor-κB (NF-κB)/Rel signaling, which controls genes involved in a broad range of biological processes, from immunity and inflammation to cell survival, in response to a wide range of stimuli [44][75]. In this context, RNF11 is involved as a negative regulator in the termination of the inflammatory response functioning.
NF-κB proteins function as dimeric transcription factors that are maintained inactive in the cytoplasm by their interaction with inhibitory IκB proteins (IκB, inhibitor of NF-κB), which mask the nuclear localization signals that are required for the nuclear import of NF-κB dimers. Signaling through a subset of receptors induces the release of the inhibitory mechanism and creates transcriptionally competent NF-κB complexes that translocate to the nucleus and induce the expression of target genes (Figure 4).
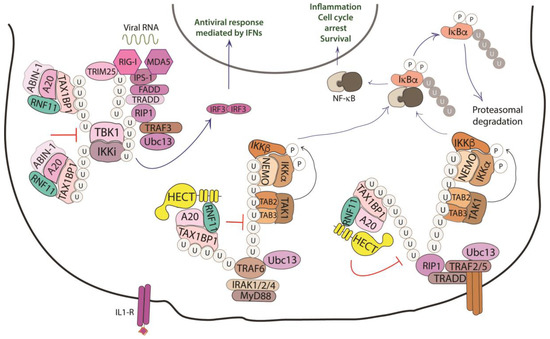
Figure 4. Role of RNF11 in the NF-κB signaling pathway. The activation of the NF-κB signaling leads to the degradation of the inhibitory IκB proteins (IκB, inhibitor of NF-κB), that mask the nuclear localization signals required for the nuclear import of NF-kB. The downstream recruitment of most of the signaling proteins is mediated by poly-ubiquitination of several components of the signal transduction machinery. E3-ligases, such as TRAF2/3/5/6 ubiquitinate the receptor interacting protein 1 (RIP1) by K11 and K63-linked ubiquitin chains. These chains serve as a scaffold for the recruitment of the linear ubiquitin chain assembly complex (LUBAC) that mediates NEMO M1-linear ubiquitination. In turn, it allows for the recruitment of TGF-β Activated Kinase 1 Binding Protein 2/3 (TAB2/3) and TAK1 that activate the IKK complex (IKK1/IKK2/NEMO). This complex triggers the ubiquitin-dependent degradation of IκB and the release of the NF-κB transcription factor. The deubiquitinating enzymes counteract the activity of the E3-ligases and play an essential function in restricting NF-kB signaling by targeting critical pathway components. In particular, the A20 ubiquitin-editing complex, comprising A20, TAX1BP1, ABIN-1, ITCH, and RNF11, catalyzes the removal of K63-linked ubiquitin chains from proteins of the signal transduction machinery, and the subsequent addition of K48-linked ubiquitin chains, promoting degradation by proteasome and termination of NF-κB signaling.
Whilst ligand-induced phosphorylation events promote binding to the activated receptors, the poly-ubiquitination of several components of the signal transduction machinery acts as a binding platform that mediates the downstream recruitment of most of the signaling proteins [45][76]. In particular, the protein family of tumor necrosis factor (TNF) receptor-associated factor (TRAF) E3-ligases mediates K63-linked ubiquitination of many of these signaling proteins. Deubiquitinating enzymes counteract the activity of the E3-ligases and play an essential function in restricting NF-kB signaling by targeting critical pathway components [46][77].
A20 is one of these DUBs, a deubiquitinating enzyme that is transcriptionally upregulated by NF-κB, which works as a ubiquitin-editing enzyme. Indeed, A20 removes Lys63-linked chains from substrates and promotes the conjugation of Lys48-linked chains that lead to proteasomal degradation of targets [47][78]. The activity of A20 is carried out together with the adaptor Tax1 Binding Protein 1 (TAX1BP1) and the E3-ligase ITCH [48][79]. TAX1BP1 functions as an adaptor that recruits ITCH to A20 and A20 to its substrates, which are the receptor interacting protein 1 (RIP1) and the E3-ligase TRAF6. Even though the precise role of ITCH in the A20 complex is still unclear, it is possible that this HECT-ligase collaborates in the ubiquitin editing function of A20 by ubiquitinating RIP1 with K48-linked ubiquitin chains [49][80]. Harhaj and colleagues have shown that RNF11 transiently associates with A20, TAX1BP1, and RIP1 following TNF stimulation and that RNF11 knockdown abrogates the assembly of the A20 complex on RIP1, which causes persistent phosphorylation of the inhibitor protein IkBα and c-Jun N-terminal kinase (JNK) [48][50][79,81]. Moreover, when RNF11 is knocked down, RIP1 and TRAF6 ubiquitination are significantly enhanced upon ligand stimulation, which indicates that RNF11 antagonizes the stimulus-dependent ubiquitination of both proteins.
The knockdown of endogenous RNF11, as well as the expression of ectopic RNF11, also inhibits IL-1 and poly (I:C)-induced NF-kB activation, which indicates that RNF11 is essential for the inactivation of key signaling molecules in both the TNFR and TLR4/IL-1R pathways. Later, publications have confirmed that the reduced expression of RNF11 results in the aberrant regulation of inflammatory signaling in neuroblastoma cells, microglia cell line, and murine cortical neurons, which suggests a tight association of RNF11 with the pathogenesis of neurodegenerative diseases [48][51][52][53][79,82,83,84].
2.3. RNF11, a Novel Regulator of E2s and E3s Activity
Numerous regulative mechanisms, extensively reviewed in the recent years [15][54][55][20,55,91], control the catalytic activity of the E3-ligases in terms of interaction with the E2, recognition of the substrate, and assembly of a specific chain. These mechanisms include post-translational modifications—mainly phosphorylations and conjugation with ubiquitin and ubiquitin-like proteins—and the noncovalent binding of proteins. The common element is represented by the fact that each enzyme is kept in a closed and inactive conformation, until a given signal triggers the release of the inhibitory state and the consequent access to the protein substrates [56][57][58][59][92,93,94,95]. Although still far from being conclusive, the experimental data accumulated so far suggest three principal roles for RNF11: as HECT substrate, HECT adaptor, and HECT inhibitor. As previously discussed, RNF11 is a substrate of HECT ligases for poly-ubiquitination reactions. The interaction with members of the NEDD4 family has been extensively analysed by several groups and the binding sites have been mapped, which confirms the WW/PPxY recognition as the main interaction mode. Accordingly, the mutation of the PY motif in RNF11 disrupted the interactions with the NEDD4 family members and stabilized RNF11 mutant protein levels, clearly indicating that at least some NEDD4 family members control RNF11 stability [12][17]. Notably, the ubiquitination of the RNF11 PY mutant is not completely abrogated and still depends on the catalytic activity of the NEDD4 family proteins. The presence of secondary contact sites [12] [17] might be important in differentiating between degradative or regulatory functions of RNF11 ubiquitination.
Data reporting the involvement of RNF11 in HECT-mediated ubiquitination pathways share the common evidence of RNF11 acting as an adaptor of HECT ligases, although this function seems to be broader and more complex than is currently known. In fact, adaptor proteins of HECT ligases typically serve the double function of acting as a bridge for the substrate recruitment and as a regulator of the catalytic activity. The adaptor function of RNF11 has been clearly demonstrated for GGA3 protein, which is ubiquitinated by ITCH in an RNF11-dependent manner and, in principle, this mechanism can be extended to all of the GGA family members, because the VHS domains embedded in these proteins are able to bind the RNF11 di-leucine motifs. The ability of RNF11 to interact with several ubiquitin binding motifs would further extend its adaptor function, thus widening the spectrum of substrates that are recruited by the HECT ligases. Moreover, the kinetic of association between RNF11 and many of its validated binding partners shows to be sensitive to the stimulation of several membrane receptors, with a transient increase in binding ability detectable at the early phases of induction. For example, binding of RNF11 to TRAF6 occurs 15–30 minutes after IL-1 stimulation, and then RNF11 is released [60][96]. A similar kinetic can be observed in the interaction with the E2 enzymes UBE2N and UbcH5, which cooperate with TRAFs and cIAPs in the TNFR signaling pathway [61][62][97,98]. Similarly, the transient complex, including RNF11, ANKRD13A, and the activated EGFR, is assembled and persists until the receptor reaches the early endosome compartment, within 15–20 minutes from plasma membrane invagination, after which RNF11 is released [35][31]. These observations suggest that the RNF11 adaptor function is strictly regulated in space and time in order to limit the interaction with substrates and/or chain elongation on the target.
RNF11 emerges as a regulator of the NEDD4-like HECT ligases. In vivo, RNF11 overexpression strongly inhibits the ITCH-mediated ubiquitination of several protein adaptors (GGA, HRS, STAM2, ANKRD13A) [13][35][18,31], as well as the ubiquitination of SMURF2 or WWP1 [63][37][38,39]. In some cases, this ability is strictly dependent on the functionality of the RING-H2 domain, while, in other cases, it is not. Moreover, there is evidence that in vitro RNF11 assembles an active E3-ligase complex with SMURF2, while the in vivo function of this complex is strongly dependent on the sorting determinants that govern the subcellular localization of the RING ligase [63][38]. This variability suggests the existence of different RNF11-mediated inhibitory mechanisms [63][38].
Whatever the mechanism, this function must require (i) the recruitment of the HECT ligase in an open conformation by disrupting the intra-molecular inhibition, (ii) the binding of a subset of substrates, and (iii) a molecular switch that allows for substrate ubiquitination by the HECT ligase only after an unidentified signal triggering the dissociation of RNF11 arrives.
To date, we can only speculate on how HECT inhibition by RNF11 can be achieved (Figure 5).
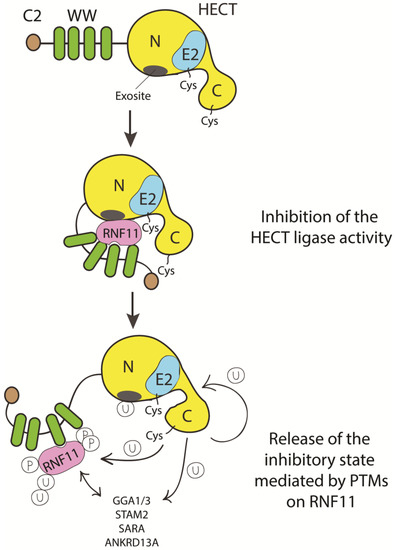
Figure 5. Cartoon representing putative inhibitory mechanisms of HECT ligases mediated by RNF11. E3-ligases of the HECT family are characterized by the presence of C2 and WW domains located upstream of the catalytic domain. RNF11 binds the WW domain containing region of all the HECT family members and these interactions are reported to inhibit HECT activity. This inhibition can result from several events. Here, we suggest that RNF11 promotes a closed conformation, which masks the E2-binding site and/or the non-covalent ubiquitin binding site (exosite) on the HECT ligase. Post-translational modifications modulated by the activation of different signaling pathways could be responsible for the conformational modifications of RNF11, allowing the recruitment of substrates on the WW domains and triggering the adaptor functions of RNF11.
