Brassinosteroids, the steroid hormones of plants, control physiological and developmental processes through its signaling pathway. The major brassinosteroid signaling network components, from the receptor to transcription factors, have been identified in the past two decades. The development of biotechnologies has driven the identification of novel brassinosteroid signaling components, even revealing several crosstalks between brassinosteroid and other plant signaling pathways.
- brassinosteroids
- signaling
- technologies
- proximity labeling
- single-molecule methods
Please note: Below is an entry draft based on your previous paper, which is wrirren tightly around the entry title. Since it may not be very comprehensive, we kindly invite you to modify it (both title and content can be replaced) according to your extensive expertise. We believe this entry would be beneficial to generate more views for your work. In addition, no worry about the entry format, we will correct it and add references after the entry is online (you can also send a word file to us, and we will help you with submitting).
1. Introduction
Brassinosteroids (BRs) are plant steroid hormones critical in numerous developmental and physiological processes, such as stem elongation, vascular differentiation, seed size, male fertility, flowering time, senescence, cell division, and resistance to biotic and abiotic stresses [1][2][1,2]. Because of the discovery of the BR receptor BR-insensitive 1 (BRI1) [3][4][3,4], several key components of the BR signaling pathway have been reported (Figure 1). BRI1 is a cell surface-localized receptor kinase [4][5][4,5]. Without BRs, BRI1 is maintained at an inactive state by its C-terminal autoinhibition [6], and by a negative regulator, BRI1 kinase inhibitor 1 (BKI1), which prevents the interaction of BRI1 with its coreceptor BRI1-associated kinase 1 (BAK1) [7][8][7,8]. In the downstream signaling pathway, two main transcription factors, BRI1 ethyl methanesulfonate (EMS) suppressor 1 (BES1) and brassinazole resistant 1 (BZR1), are phosphorylated by BR-insensitive 2 (BIN2), and some members of 14-3-3 proteins can bind and retain the phosphorylated BES1 and BZR1 in the cytoplasm [9][10][11][12][9,10,11,12]. BR binding to the island domain of BRI1 induces a conformational change of the ectodomain [13][14][13,14], which activates the intracellular domain, releases the autoinhibition of BRI1 C-terminus, and phosphorylates BKI1 [6][8][15][6,8,15]. The phosphorylated BKI1 dissociates from the plasma membrane to interact with 14-3-3λ and κ, and competitively inhibits the binding of 14-3-3λ and κ with BES1/BZR1, leading to BR-regulated gene expression [15]. At the same time, the activated BRI1 interacts with and transphosphorylates its coreceptor BAK1 to recruit positive regulators, such as BR signaling kinases (BSKs) and constitutive differential growth 1 (CDG1), and phosphorylates them [16][17][16,17]. Phosphorylated BSKs and CDG1 then activate phosphatase BRI1-suppressor 1 (BSU1) and its homologs BSU1 like (BSLs) to inactivate BIN2 [16][17][16,17]. Subsequently, protein phosphatase 2A (PP2A) dephosphorylates BES1/BZR1, leading to the expression of the BR-responsive genes [18][19][18,19]. Several reviews have already summarized the most recent discoveries in the BR signaling pathway [2][20][2,20]. Here, we introduce several technologies and methodologies that have been applied to the study of BR signal transduction and have led to discoveries unattainable by conventional methods. Although our focus is on the BR pathway, the applications of each method can provide inspirations for studies on other plant signaling pathways.
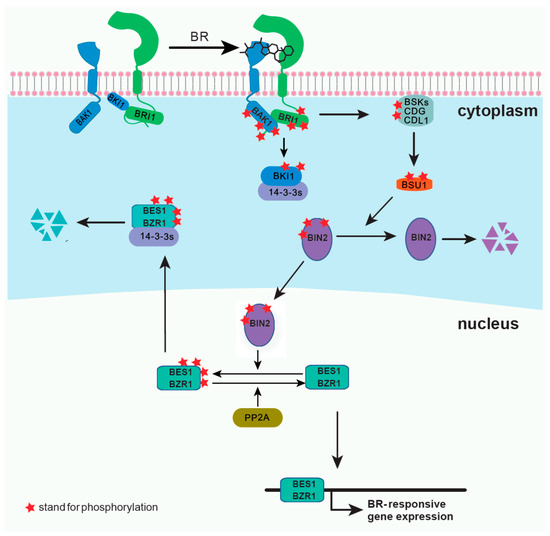
Figure 1.
The development of the understanding of the BR signaling pathway has progressed with the advancements in biotechnology. Molecular genetics provides the fundamentals for the study of signaling pathways. In particular, the combination of forward and reverse genetics facilitated the identification of BR signaling components. Here, we summarize traditional and new methods used. Conventional methods, including EMS mutant screening, activation tagging, yeast two-hybrid (Y2H) screening, two-dimensional (2D) difference gel electrophoresis (DIGE), and liquid chromatography (LC)–tandem mass spectrometry (MS/MS), the applications of which have laid the foundation of our current understanding of the BR signaling pathway. New technologies, such as bioinformatics, clustered regularly interspaced short palindromic repeats (CRISPR) system, proximity labeling (PL), and single-molecule technology, have considerably enhanced this understanding.
2. Identification of New Components of BR Signaling
2.1. Forward Genetics
Forward genetics is the screening of phenotypes of interest under different backgrounds and conditions to identify the gene that causes a certain phenotype. It laid the foundation for our understanding of the BR signaling pathway in the early days. The application of forward genetic screening usually begins with naturally or artificially generated mutations.
2.2. Reverse Genetics
In contrast to forward genetics, reverse genetics starts from molecular engineering of a gene to search for alterations of phenotypes. Genome sequencing enables easy characterization of the gene sequence. Several popular methods directly identify signaling components: Y2H, two-dimensional polyacrylamide gel electrophoresis, LC–MS/MS, bioinformatic prediction, and CRISPR.
3. Examination of the Dynamic Regulation Mechanism of BR Signaling Components
After the main components of BR signaling are identified, the regulation mechanism between them is examined, including interactions, structure analysis, phosphorylation, acetylation, and polyubiquitination. However, these mechanisms are in a quiescent condition. Proximity labeling (PL) is a powerful new technique for studying protein–protein interactions (PPIs). In recent years, the dynamic interaction has also been examined using single-molecule technology.
3.1. Plant Hormone Signaling Pathway Studied Using PL
PPIs are the basic elements of signaling pathways. Currently, the gold standards for studying PPIs are Y2H, co-immunoprecipitation, pulldown assays, and in vivo Förster-type resonance energy transfer (FRET) or bimolecular fluorescence complementation (BiFC) assays. However, each of these has its own limitations. Y2H, co-immunoprecipitation, and pulldown assays can be used to explore potential unknown interaction partners of a bait protein. However, they usually suffer from false negatives and false positives. Furthermore, they fail to detect interactions that are weak or appear only transiently. Moreover, for membrane lipid-assisted PPIs, the trace of interactions can easily be lost during the harsh lysing and affinity pulldown processes. In vivo FRET or BiFC assays can, in principle, capture transient PPIs, but these methods are in the binary candidate format and require fluorescent labeling. Therefore, the successful execution of the assays depends on the transfection efficiency, and the assays are prone to disruption by the native autofluorescence background of live cells, especially plant cells. PL has been a perfect complement for the conventional PPI assays. PL uses engineered ligase or peroxidase to cast specific tags, usually biotin, on certain residues on proteins appearing in the proximity of the original bait protein. The interactome information can be analyzed from the cell lysate with affinity chromatography–MS/MS.
The BR signaling pathway was investigated through TurboID-mediated PL mapping of BIN2 proteomics [21], and 280 proteins were identified in the BIN2-YFP-TbID run but not in the control set. The crossover of BIN2 from the BR network to other signaling pathways was also observed. The auxin transporter PIN3 was identified through PL–MS, explaining the reason that BR modulates polar auxin transport [22][23]. Phototropism-related genes, phototropin 1 (PHOT1), nonphototropic hypocotyl 3 (NPH3), and an NPH3 family protein were also found. The findings suggest how BR mediates plant development while mitigating various other pathways through the negative regulatory factor BIN2.
The BR signaling pathway was investigated through TurboID-mediated PL mapping of BIN2 proteomics [67], and 280 proteins were identified in the BIN2-YFP-TbID run but not in the control set. The crossover of BIN2 from the BR network to other signaling pathways was also observed. The auxin transporter PIN3 was identified through PL–MS, explaining the reason that BR modulates polar auxin transport [68,69]. Phototropism-related genes, phototropin 1 (PHOT1), nonphototropic hypocotyl 3 (NPH3), and an NPH3 family protein were also found. The findings suggest how BR mediates plant development while mitigating various other pathways through the negative regulatory factor BIN2.
3.2. Single-Molecule Technologies for Studying BR System
Single-molecule methods facilitate the observation of the dynamic interaction processes of target molecules. Compared with the aforementioned conventional techniques, which can identify new components of signaling but provide limited quantitative and dynamic data, single-molecule methods study biological macromolecules one-by-one in real-time and provide valuable insights into the fundamental biochemical and biophysical properties of the target biological system. However, only a few studies have used these methods to examine BR signaling transduction. For example, Wang et al. used variable-angle total internal reflection fluorescence microscopy (VA-TIRFM) and fluorescence correlation spectroscopy/fluorescence cross-correlation spectroscopy (FCS/FCCS) [24][25][70,71], Song et al. used colocalization single-molecule spectroscopy (CoSMoS) based on total internal reflection fluorescence microscopy (TIRFM) [26][27][72,73], and Hink et al. used the photon counting histogram (PCH) model and FCCS [25][28][71,74]. These studies have provided unique information on the BR system and demonstrated how single-molecule methods are powerful and promising tools for studying fundamental questions in plant science.
4. Concluding Remarks
Various methodologies can be applied to facilitate the identification of new BR members and foster our comprehension of BR signaling and other signaling networks. In this review, we introduce several recent developments of technologies and illustrate how they contribute to our current understanding of the phytohormone BR. The method used in BR signaling is also available for other signaling pathways, particularly PL and single-molecule technologies. Notably, not many applications of these novel methodologies exist in the field of BR research, indicating that the usage of these methods is still at its budding stage, and many opportunities await. We believe that these methods will develop into powerful plant science research tools.
In addition, there are still many unanswered questions in BR signaling pathway research such as how the tissue specific development is regulated and whether non-coding RNA functions in BR signaling regulation. Besides phosphorylation and acetylation, does another modification exist and how does it regulate the BR signal? How do the redundant genes work? How does the BR signal affect modifications of the whole plants? Therefore, we still need to develop new methods for different conditions in BR signaling research, such as tissue specific signal-cell sequencing, next generation sequencing, and proteomics of post-translational modifications. Moreover, almost all of these methods are based on bioinformatics. Furthermore, comparative studies of BR signal pathway across different plant species will lead to a better understanding of the evolution and better application of BR signal pathway.
