Various deep-learning (DL) methods that utilize a combination of omics data and imaging data have been applied to the diagnosis, prognosis, and treatment options of clinical COVID-19 patients. Even with the emerging deep-learning methods, human intervention is still essential in the clinical diagnosis and treatment of COVID-19 patients.
- artificial intelligence
- deep learning
- multi-modal learning
1. Deep Learning for Diagnosis of COVID-19
Various deep-learning (DL) methods that utilize a combination of omics data and imaging data have been applied to the diagnosis, prognosis, and treatment options of clinical COVID-19 patients. However, even with the emerging deep-learning methods, human intervention is still essential in the clinical diagnosis and treatment of COVID-19 patients. Therefore, the goal of DL is not to surpass or replace humans, but rather to provide decision-support tools to help researchers studying COVID-19 and health professionals in the clinical management of COVID-19 patients.
[1]
Figure 1
[4]
[5]
[6]
[7]
[8]
[9]
[10]
[11]
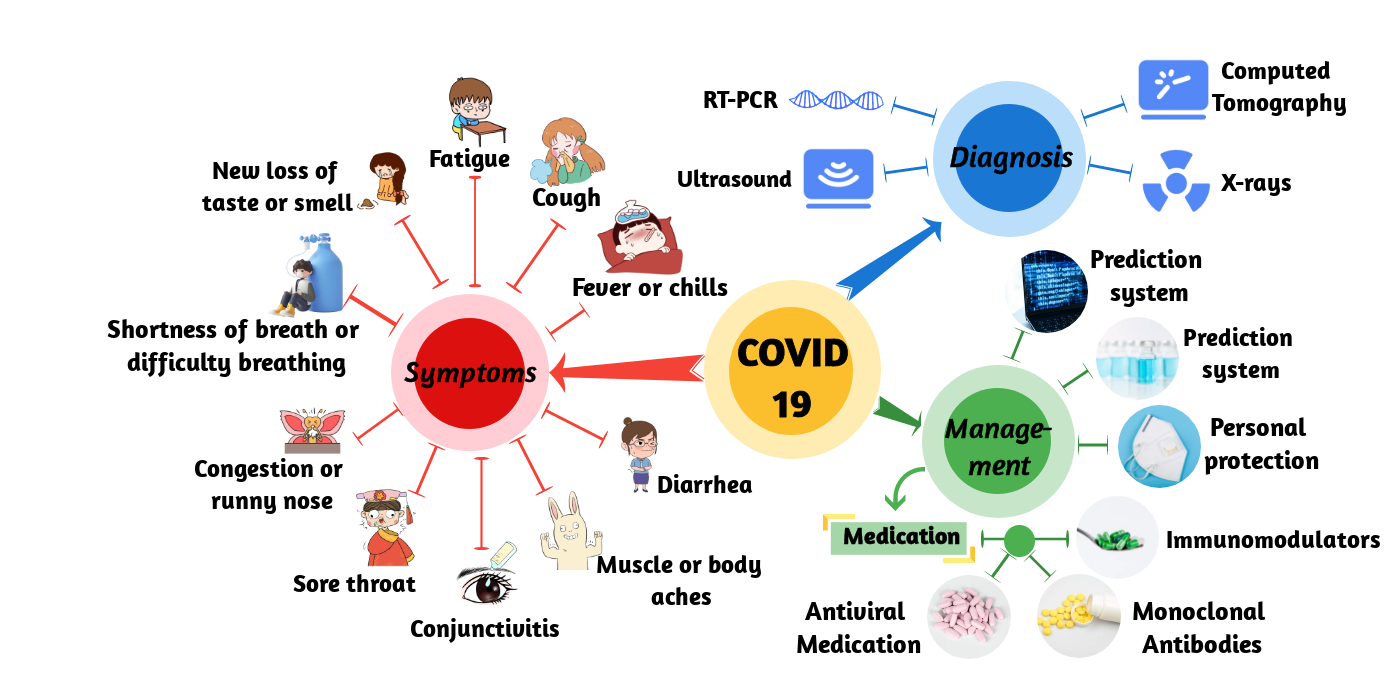
Figure 1. Schematic of COVID-19. It shows the symptoms, diagnosis, and management of COVID-19.
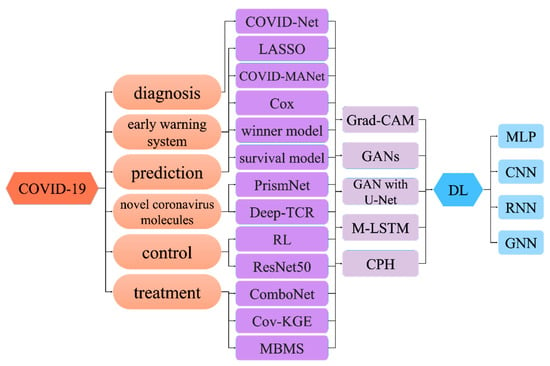
2. Deep Learning for COVID-19 Early Warning System
3. Deep Learning for COVID-19 Prediction
4. Deep Learning for Novel Coronavirus Molecules
5. Deep Learning for COVID-19 Control
6. Deep Learning for COVID-19 Treatment
References
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in different types of clinical specimens. Jama 2020, 323, 1843–1844.
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720.
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506.
- Liu, L.; Gao, J.-Y.; Hu, W.-M.; Zhang, X.-X.; Guo, L.; Liu, C.-Q.; Tang, Y.-W.; Lang, C.-H.; Mou, F.-Z.; Yi, Z.-J.; et al. Clinical characteristics of 51 patients discharged from hospital with COVID-19 in chongqing, China. medRxiv 2020.
- Wang, L.; Lin, Z.Q.; Wong, A. Covid-net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest x-ray images. Sci. Rep. 2020, 10, 19549.
- Hu, Q.; Gois, F.N.B.; Costa, R.; Zhang, L.; Yin, L.; Magaia, N.; de Albuquerque, V.H.C. Explainable artificial intelligence-based edge fuzzy images for COVID-19 detection and identification. Appl. Soft Comput. 2022, 123, 108966.
- Chouhan, V.; Singh, S.K.; Khamparia, A.; Gupta, D.; Tiwari, P.; Moreira, C.; Damaševičius, R.; de Albuquerque, V.H.C. A novel transfer learning based approach for pneumonia detection in chest x-ray images. Appl. Sci. 2020, 10, 559.
- Afshar, P.; Heidarian, S.; Naderkhani, F.; Oikonomou, A.; Plataniotis, K.N.; Mohammadi, A. Covid-caps: A capsule network-based framework for identification of COVID-19 cases from x-ray images. Pattern Recognit. Lett. 2020, 138, 638–643.
- Li, Z.; Liu, F.; Yang, W.; Peng, S.; Zhou, J. A survey of convolutional neural networks: Analysis, applications, and prospects. IEEE Trans. Neural Netw. Learn. Syst. 2022, 33, 6999–7019.
- Bayoudh, K.; Hamdaoui, F.; Mtibaa, A. Hybrid-covid: A novel hybrid 2d/3d cnn based on cross-domain adaptation approach for COVID-19 screening from chest x-ray images. Phys. Eng. Sci. Med. 2020, 43, 1415–1431.
- Zhang, J.; Yu, L.; Chen, D.; Pan, W.; Shi, C.; Niu, Y.; Yao, X.; Xu, X.; Cheng, Y. Dense gan and multi-layer attention based lesion segmentation method for COVID-19 ct images. Biomed. Signal Process. Control 2021, 69, 102901.
- Shi, W.; Tong, L.; Zhu, Y.; Wang, M.D. COVID-19 automatic diagnosis with radiographic imaging: Explainable attention transfer deep neural networks. IEEE J. Biomed. Health Inform. 2021, 25, 2376–2387.
- Diaz-Escobar, J.; Ordóñez-Guillén, N.E.; Villarreal-Reyes, S.; Galaviz-Mosqueda, A.; Kober, V.; Rivera-Rodriguez, R.; Rizk, J.E.L. Deep-learning based detection of COVID-19 using lung ultrasound imagery. PLoS ONE 2021, 16, e0255886.
- Fang, C.; Bai, S.; Chen, Q.; Zhou, Y.; Xia, L.; Qin, L.; Gong, S.; Xie, X.; Zhou, C.; Tu, D.; et al. Deep learning for predicting COVID-19 malignant progression. Med. Image Anal. 2021, 72, 102096.
- Näppi, J.J.; Uemura, T.; Watari, C.; Hironaka, T.; Kamiya, T.; Yoshida, H. U-survival for prognostic prediction of disease progression and mortality of patients with COVID-19. Sci. Rep. 2021, 11, 9263.
- Sun, C.; Hong, S.; Song, M.; Li, H.; Wang, Z. Predicting COVID-19 disease progression and patient outcomes based on temporal deep learning. BMC Med. Inform. Decis. Mak. 2021, 21, 1–16.
- Uemura, T.; Näppi, J.J.; Watari, C.; Hironaka, T.; Kamiya, T.; Yoshida, H. Weakly unsupervised conditional generative adversarial network for image-based prognostic prediction for COVID-19 patients based on chest ct. Med. Image Anal. 2021, 73, 102159.
- Vaid, S.; Kalantar, R.; Bhandari, M. Deep learning COVID-19 detection bias: Accuracy through artificial intelligence. Int. Orthop. 2020, 44, 1539–1542.
- Ikemura, K.; Bellin, E.; Yagi, Y.; Billett, H.; Saada, M.; Simone, K.; Stahl, L.; Szymanski, J.; Goldstein, D.Y.; Gil, M.R. Using automated machine learning to predict the mortality of patients with covid-19: Prediction model development study. J. Med. Internet Res. 2021, 23, e23458.
- Meng, L.; Dong, D.; Li, L.; Niu, M.; Bai, Y.; Wang, M.; Qiu, X.; Zha, Y.; Tian, J. A deep learning prognosis model help alert for COVID-19 patients at high-risk of death: A multi-center study. IEEE J. Biomed. Health Inform. 2020, 24, 3576–3584.
- Suppakitjanusant, P.; Sungkanuparph, S.; Wongsinin, T.; Virapongsiri, S.; Kasemkosin, N.; Chailurkit, L.; Ongphiphadhanakul, B. Identifying individuals with recent COVID-19 through voice classification using deep learning. Sci. Rep. 2021, 11, 1–7.
- Liang, W.; Yao, J.; Chen, A.; Lv, Q.; Zanin, M.; Liu, J.; Wong, S.; Li, Y.; Lu, J.; Liang, H.; et al. Early triage of critically ill COVID-19 patients using deep learning. Nat. Commun. 2020, 11, 1–7.
- Dong, Y.M.; Sun, J.; Li, Y.X.; Chen, Q.; Liu, Q.Q.; Sun, Z.; Pang, R.; Chen, F.; Xu, B.Y.; Manyande, A.; et al. Development and validation of a nomogram for assessing survival in patients with COVID-19 pneumonia. Clin. Infect. Dis. 2021, 72, 652–660.
- Gao, J.; Sharma, R.; Qian, C.; Glass, L.M.; Spaeder, J.; Romberg, J.; Sun, J.; Xiao, C. Stan: Spatio-temporal attention network for pandemic prediction using real-world evidence. J. Am. Med. Inform. Assoc. 2021, 28, 733–743.
- Dairi, A.; Harrou, F.; Zeroual, A.; Hittawe, M.M.; Sun, Y. Comparative study of machine learning methods for COVID-19 transmission forecasting. J. Biomed. Inform. 2021, 118, 103791.
- Liao, Z.; Song, Y.; Ren, S.; Song, X.; Fan, X.; Liao, Z. Voc-dl: Deep learning prediction model for COVID-19 based on voc virus variants. Comput. Methods Programs Biomed. 2022, 224, 106981.
- Mary, S.R.; Kumar, V.; Venkatesan, K.J.P.; Kumar, R.S.; Jagini, N.P.; Srinivas, A. Vulture-based adaboost-feedforward neural frame work for COVID-19 prediction and severity analysis system. Interdiscip Sci. 2022, 14, 582–595.
- Mansour, R.F.; Escorcia-Gutierrez, J.; Gamarra, M.; Gupta, D.; Castillo, O.; Kumar, S. Unsupervised deep learning based variational autoencoder model for COVID-19 diagnosis and classification. Pattern Recognit. Lett. 2021, 151, 267–274.
- Liao, Z.; Lan, P.; Fan, X.; Kelly, B.; Innes, A.; Liao, Z. Sirvd-dl: A COVID-19 deep learning prediction model based on time-dependent sirvd. Comput. Biol. Med. 2021, 138, 104868.
- Kafieh, R.; Arian, R.; Saeedizadeh, N.; Amini, Z.; Serej, N.D.; Minaee, S.; Yadav, S.K.; Vaezi, A.; Rezaei, N.; Javanmard, S.H. COVID-19 in iran: Forecasting pandemic using deep learning. Comput. Math. Methods Med. 2021, 2021, 1–16.
- Taz, T.A.; Ahmed, K.; Paul, B.K.; Kawsar, M.; Aktar, N.; Mahmud, S.M.H.; Moni, M.A. Network-based identification genetic effect of SARS-CoV-2 infections to idiopathic pulmonary fibrosis (ipf) patients. Brief. Bioinform. 2020, 22, 1254–1266.
- Mahmud, S.M.H.; Al-Mustanjid, M.; Akter, F.; Rahman, M.S.; Ahmed, K.; Rahman, M.H.; Chen, W.; Moni, M.A. Bioinformatics and system biology approach to identify the influences of SARS-CoV-2 infections to idiopathic pulmonary fibrosis and chronic obstructive pulmonary disease patients. Brief. Bioinform. 2021, 22, bbab115.
- Sun, L.; Li, P.; Ju, X.; Rao, J.; Huang, W.; Ren, L.; Zhang, S.; Xiong, T.; Xu, K.; Zhou, X.; et al. In vivo structural characterization of the SARS-CoV-2 rna genome identifies host proteins vulnerable to repurposed drugs. Cell 2021, 184, 1865–1883.e1820.
- Sidhom, J.W.; Baras, A.S. Deep learning identifies antigenic determinants of severe SARS-CoV-2 infection within t-cell repertoires. Sci. Rep. 2021, 11, 14275.
- Yang, D.; Yurtsever, E.; Renganathan, V.; Redmill, K.A.; Özgüner, Ü. A vision-based social distancing and critical density detection system for COVID-19. Sensors 2021, 21, 4608.
- Zhang, T.; Li, J. Understanding and predicting the spatio-temporal spread of COVID-19 via integrating diffusive graph embedding and compartmental models. Trans GIS 2021, 25, 3025–3047.
- Jin, W.; Stokes, J.M.; Eastman, R.T.; Itkin, Z.; Zakharov, A.V.; Collins, J.J.; Jaakkola, T.S.; Barzilay, R. Deep learning identifies synergistic drug combinations for treating COVID-19. Proc. Natl. Acad. Sci. USA 2021, 118, e2105070118.
- Ottakath, N.; Elharrouss, O.; Almaadeed, N.; Al-Maadeed, S.; Mohamed, A.; Khattab, T.; Abualsaud, K. Vidmask dataset for face mask detection with social distance measurement. Displays 2022, 73, 102235.
- Siah, C.R.; Lau, S.T.; Tng, S.S.; Chua, C.H.M. Using infrared imaging and deep learning in fit-checking of respiratory protective devices among healthcare professionals. J. Nurs. Sch. 2022, 54, 345–354.
- Sethi, S.; Kathuria, M.; Kaushik, T. Face mask detection using deep learning: An approach to reduce risk of coronavirus spread. J. Biomed. Inform. 2021, 120, 103848.
- Zeng, Y.; Chen, X.; Luo, Y.; Li, X.; Peng, D. Deep drug-target binding affinity prediction with multiple attention blocks. Brief. Bioinform. 2021, 22, bbab117.
- Nguyen, D.D.; Gao, K.; Chen, J.; Wang, R.; Wei, G.W. Unveiling the molecular mechanism of SARS-CoV-2 main protease inhibition from 137 crystal structures using algebraic topology and deep learning. Chem. Sci. 2020, 11, 12036–12046.
- Zeng, X.; Song, X.; Ma, T.; Pan, X.; Zhou, Y.; Hou, Y.; Zhang, Z.; Li, K.; Karypis, G.; Cheng, F. Repurpose open data to discover therapeutics for COVID-19 using deep learning. J. Proteome Res. 2020, 19, 4624–4636.
- Roh, H.; Shin, S.; Han, J.; Lim, S. A deep learning-based medication behavior monitoring system. Math. Biosci. Eng. 2021, 18, 1513–1528.
- Santos, M.A.G.; Munoz, R.; Olivares, R.; Filho, P.P.R.; Ser, J.D.; de Albuquerque, V.H.C. Online heart monitoring systems on the internet of health things environments: A survey, a reference model and an outlook. Inf. Fusion 2020, 53, 222–239.
- Parah, S.A.; Kaw, J.A.; Bellavista, P.; Loan, N.A.; Bhat, G.M.; Muhammad, K.; de Albuquerque, V.H.C. Efficient security and authentication for edge-based internet of medical things. IEEE Internet Things J. 2021, 8, 15652–15662.
