Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Rebecca Wingert.
Nephrons are the functional units which comprise the kidney. Each nephron contains a number of physiologically unique populations of specialized epithelial cells that are organized into discrete domains known as segments.
- nephron
- development
- zebrafish
1. Introduction
1.1. Essential Functions of the Kidney
The kidney performs various vital functions in the body, such as removing metabolic waste products, monitoring blood pressure, secreting hormones and maintaining pH, electrolyte and water balance [1]. Nephron functional units in the kidney consist of subdomains, or segments, that are dedicated to particular physiological tasks (Figure 1) [2,3][2][3]. These segment regions include the renal corpuscle that filters the blood; tubule segments that modify the filtrate, including the proximal tubule, loop of Henle, distal tubule and connecting tubule; and lastly, the collecting duct, which performs the final modifications on urine and conveys it out of the kidney [4,5,6,7,8,9,10,11,12][4][5][6][7][8][9][10][11][12]. Within the renal corpuscle, the podocytes secrete collagen and growth factors to help maintain glomerular basement membrane and endothelial cell fenestration, respectively [4,5][4][5]. Additionally, the ultrastructural features of the podocytes are crucial components in establishing glomerular filtration [4,5][4][5]. The proximal tubule is important for reabsorbing nutrients, electrolytes and water: in total, it reabsorbs about two thirds of most water and important electrolytes, such as Na+, Cl− and HCO3−, as well as 99.8% of glucose and amino acids filtered by the human kidney [6,7][6][7]. The loop of Henle is further divided into the descending thin limb (DTL), the ascending thin limb (ATL) and the thick ascending limb (TAL), which together play critical roles in water homeostasis [8]. This is achieved by the expression of the Na+/K+/2Cl− cotransporter in the TAL, which helps the reabsorption of 25% of filtered Na+ [9]. Additionally, the TAL also reabsorbs filtered Ca2+ and Mg2+ [9]. The distal tubule is important in regulating electrolytes such as Na+, K+ and Ca2+ and pH levels [10]. The distal tubule reabsorbs approximately 5–10% of filtered Na+, as well as 7–10% of filtered calcium [10]. The collecting duct originates from the ureteric bud, and is essential in regulating water and electrolytes such as Na+ and Cl− [11,12][11][12].
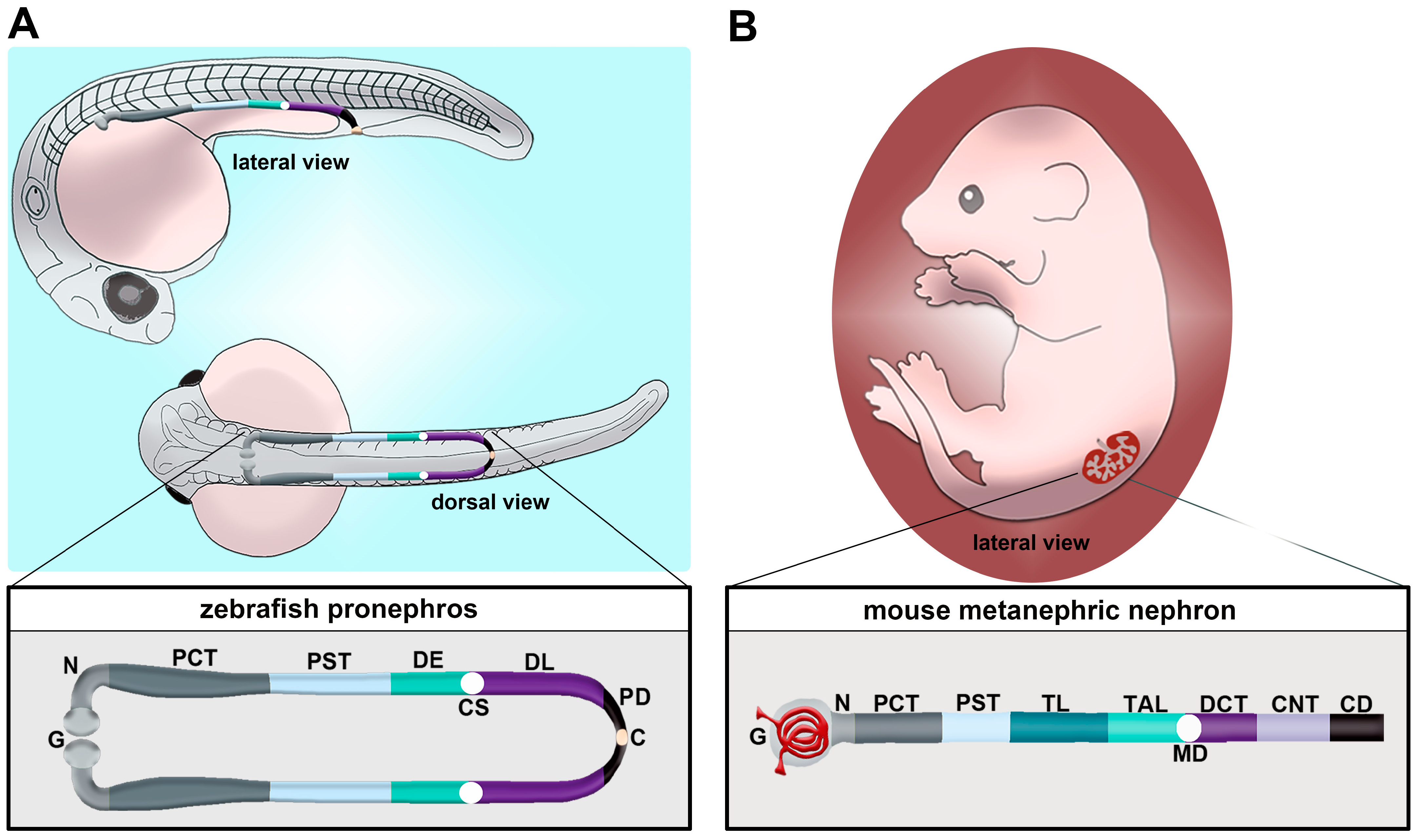
Figure 1. Comparison of nephron segment composition between the zebrafish and the mouse. (A) The zebrafish embryo forms a pronephros that consists of two nephrons located on either side of the midline, which form by the 24 h post fertilization (hpf) stage and quickly undergo morphogenesis events that enable them to begin blood filtration by approximately 48 hpf. (B) Nephron composition in the mouse metanephros; note that the nephron is drawn here in a linear configuration, but it would display a folded/convoluted anatomical configuration within the native kidney. There is a striking conservation of proximal and distal segments, but the thin limb is one notable distinction between these nephron forms. Abbreviations are as follows: P = podocytes; N = neck; PCT = proximal convoluted tubule; PST = proximal straight tubule; DE = distal early; CS = Corpuscle of Stannius; DL = distal late; CD = collecting duct; TL = thin limb; TAL = thick ascending limb; MD = macula densa; DCT = distal convoluted tubule; CNT = connecting tubule.
Given the crucial tasks accomplished by kidney nephrons, their proper formation is vital for normal renal function [13,14,15][13][14][15]. Alterations in the number and composition of nephrons have significant ramifications for kidney health, and can lead to congenital anomalies of the kidney and urinary tract (CAKUT) [16,17,18,19,20,21,22][16][17][18][19][20][21][22]. Understanding the mechanisms of nephrogenesis has many potential applications to treat both inherited and acquired renal diseases [23,24,25][23][24][25].
1.2. Vertebrate Kidney Forms Are Comprised of Nephrons
Several versions of the kidney organ are made and degraded during vertebrate ontogeny [26,27][26][27]. They develop in a prototypical sequence: the first form is termed the pronephros, the second form is termed the mesonephros and the third possible form is termed the metanephros. This last form is typically made in reptiles, birds and mammals. Across all forms, the nephron is the common structural and functional unit. In general, each version of the kidney in a given species exhibits increasing complexity with regard to the absolute number and arrangement of its nephrons. There is broad conservation of nephron segment composition. However, there are notable variations across the phylogenetic spectrum, such as the aglomerular nephrons in some species, and differences in whether nephrons possess a loop of Henle segment [23,28,29,30,31,32,33,34][23][28][29][30][31][32][33][34].
2. Using the Zebrafish to Study Nephron Development
2.1. Zebrafish Pronephros Composition and Function
Unlike mammals, which manifest the pronephros, mesonephros and metanephros during development, the zebrafish only manifests the pronephros and the mesonephros [37][35]. Each of these kidney structures originates from populations of mesodermally derived renal progenitors. The zebrafish pronephros is extremely simple, being comprised of two nephrons that form rapidly and function throughout the first several weeks of life [53,54,55,56,57,58,59,60,61,62,63,64][36][37][38][39][40][41][42][43][44][45][46][47]. Key transcription factors such as Pax2a, Pax8, Hand1 and Osr1 have critical roles in the specification of the renal progenitor’s fate [55,65,66,67,68,69,70,71,72][38][48][49][50][51][52][53][54][55]. Once this mesenchymal identity is set, which is thought to transpire from the tailbud stage or 10 h post fertilization (hpf) through to 24 hpf, the renal progenitors undergo events that transition them to form tubules with defined proximal and distal epithelial cell identities [73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90][56][57][58][59][60][61][62][63][64][65][66][67][68][69][70][71][72][73]. The zebrafish segments include the neck (N), proximal convoluted tubule (PCT), proximal straight tubule (PST), distal early (DE) tubule and distal late (DL) tubule (Figure 1A) [73][56]. Compared to the typical mammalian nephron, the loop of Henle is missing (Figure 1B) [73][56]. At approximately 48 hpf, the pronephros will become functional and begin to filter the circulation [60][43]. Between 24 hpf and 48 hpf, there are several morphogenesis events which happen to create this functioning pronephros [37][35]. For example, podocyte precursors migrate to the midline and recruit blood vessels, forming the glomerulus. The tubular segments continue to differentiate, showing the expression of additional solute transporter genes that impart unique physiological capabilities to each segment. Combined with this is the growth of the renal tubule and coiling event of the PCT.2.2. Zebrafish Mesonephros Composition and Function: Spotlight on Renal Regeneration Studies
The adult form of the zebrafish kidney, also known as the mesonephros, develops beginning around 12–14 days post fertilization (dpf) [37,91,92][35][74][75]. During this process, more nephrons are created and joined with the existing pronephros tubules, forming a small collection of interconnected nephrons that drain via a pair of renal collecting ducts [37,91,92][35][74][75]. Eventually, this process generates between 300–500 nephrons [91,92][74][75]. The zebrafish body size positively correlates with the number of mesonephric nephrons [91,92][74][75]. After complete mesonephros generation, the zebrafish possesses more complex nephron branching, yet the segmental composition remains similar to that of the pronephros [92][75]. The adult mesonephros is ultimately located at the dorsal body wall surrounded by connective tissues. In the zebrafish, the mesonephros contains a head region, a saddle and a tail region. Additionally, based on the density of nephrons throughout the mesonephros, there are four further subdivisions: anterior nephron-dense region (ANDR), medial nephron-sparse region (MNSR), medial nephron-dense region (MNDR) and posterior nephron-sparse region (PNSR). Compared to mammalian kidneys, which cannot regenerate nephrons, in the zebrafish kidney, zebrafish keep adding nephrons to their kidney throughout their lifetime. In addition to nephron addition throughout development, the mesonephros can also regenerate damage to existing nephrons after injury [91,92,93,94,95,96,97,98,99,100,101,102][74][75][76][77][78][79][80][81][82][83][84][85].3. Molecular Genetic Toolkit in the Zebrafish Animal Model
3.1. Zebrafish as a Model for Developmental Biology and Biomedical Research
The zebrafish offers a simple, yet effective, animal model to study developmental biology and biomedical research [103,104,105,106,107,108,109][86][87][88][89][90][91][92]. There are multiple traits that facilitate developmental research. First, the zebrafish has a simple architecture: at its embryonic stage, the embryonic kidney only comprises two nephrons [53][36]. Much due to their simplicity, zebrafish carry high similarity with mammalian kidneys: 70% of human genes share at least one homolog with the zebrafish [106][89]. The zebrafish kidney also shares most of the kidney segments with the mammalian kidney, with the exception of the mammalian loop of Henle [73][56]. Furthermore, zebrafish development occurs ex utero, and embryonic zebrafish are optically transparent, allowing for live imaging and visualization of organs. Next, zebrafish have high regeneration ability: new kidney nephrons keep being added to the developing kidney throughout the lifetime or after injury. Meanwhile, mammalian kidneys cannot regenerate or produce more nephrons after they are destroyed. Additionally, zebrafish demonstrate fast organogenesis, with the vital organs such as the kidney, heart and eyes available as early as 24 hpf. At 48 hpf, the kidney is fully functional, and all common organs are visible at 120–144 hpf. Coupled with this is high fecundity: zebrafish in a good laboratory environment can breed all year round, and each week they can breed up to 300 eggs.3.2. Forward Genetics: From Random Mutagenesis Screens to Chemical Screens
In forward genetic approaches, the screening of animal populations that have random mutations in the genome can uncover the novel importance of genes in biological pathways. To start this process, the first step is creating mutagenic lesions on the genome that are heritable and phenotypically distinguishable. There are several types of random mutagenesis which have been studied. Early work used gamma irradiation to create breaks in the chromosome to create mutant phenotypes. This method has a weakness as it causes large deletions, translocations or chromosomal aberrations that complicate the process to specify the genes responsible for the mutant phenotype. Subsequently, alkylating agents were used, such as ethylnitrosourea (ENU). In this process, chemical mutagens were exposed to adult males, causing random point mutations in their DNA [53][36]. The adult males were then bred with WT females, generating heterozygous F1. To expand heterozygous populations, F1 heterozygotes were then outcrossed again with WT to generate F2 heterozygous generation. F2 heterozygotes were then incrossed with each other to generate the desired homozygous mutants. This method provides more advantages over gamma irradiation, such as high mutagenic loads in the zebrafish genome, as well as the induced phenotypes being specifically linked to one gene. A major drawback of this method is that it is difficult to identify specific mutations responsible for the induced mutant phenotype. To overcome this challenge, several groups have introduced replication-deficient retroviruses or transposons as mutagens, which can insert into the genome to cause mutations. In this method, the mutagenic agent is inserted into the 1-cell stage if it is a transposon, or the 1000-cell stage embryos if it is a retrovirus. However, the overall mutagenic frequency from retroviruses and transposons has been estimated to be much lower than ENU, thus requiring a significantly more bountiful library of mutagenized fish. Both random mutagenesis and insertional mutagenesis methods have led to the generation of mutant collections with defects in kidney development [53,77,90,110,111,112,113][36][60][73][93][94][95][96]. Within the zebrafish pronephros, examples of aberrant morphological defects that could be attributed to reduced kidney function include pericardial edema, kidney cysts, hydrocephalus of the brain or body curvature [53,77][36][60]. Since the early live screens, scientists have expanded the methods of identifying mutant features. One simple way of phenotypic screening is by using a molecular marker(s) to visualize a particular cell or tissue type, using techniques such as whole mount in situ hybridization (WISH) or immunostaining [111][94]. The use of a tissue-specific transgenic line marked by fluorescent protein expression can alleviate the hands-on effort and sample fixation issue caused by in situ hybridization or immunostaining. In addition, restriction enzymes could offer a simple and useful tool to detect mutation. If the mutation affects a particular site on the DNA that a restriction enzyme has been known to affect, such as the removal of a restriction enzyme site from WT embryos, a restriction enzyme digest assay could be used to distinguish between WT and mutant embryos. Lastly, DNA extracted from embryos from mutant lines could be used for PCR and sent for sequencing, allowing for robust and direct identification of the genetic lesion.3.3. Reverse Genetics: Loss-of-Function Methods Using Morpholinos and Genome Editing
Loss-of-function studies have been performed using various approaches, including morpholino oligos, CRISPR/Cas9 system, TALENs and ZFNs [46,108][91][97]. In comparison, techniques with RNA interference (RNAi) have proven to be problematic in zebrafish [114][98]. Morpholinos are a short antisense oligonucleotide, up to 25 basepairs (bps) long, designed to target the processed mRNA or pre-mRNA transcript of the animal [115][99]. Morpholino oligos that target the ATG start site block translation, while morpholinos that target the splice donor or acceptor site of a given exon–intron boundary can disrupt the spliceosome [115][99]. This can lead to numerous outcomes, such as introns being present after RNA processing. In such a case, a splice site morpholino might cause a frameshift mutation, resulting in a premature stop codon during translation, causing a truncated protein that will later be degraded. A benefit of splice-site morpholino over ATG-site morpholino is that RNA transcripts can be reverse transcribed into DNA, and the presence of the mis-spliced transcripts can be detected using RT-PCR and sequencing [115,116][99][100]. In the zebrafish model, morpholinos are typically delivered to the 1-cell-stage embryo via microinjection, and effects last for up to several days (1–3, or more), allowing for an easy model to study development (Figure 2A) [115][99].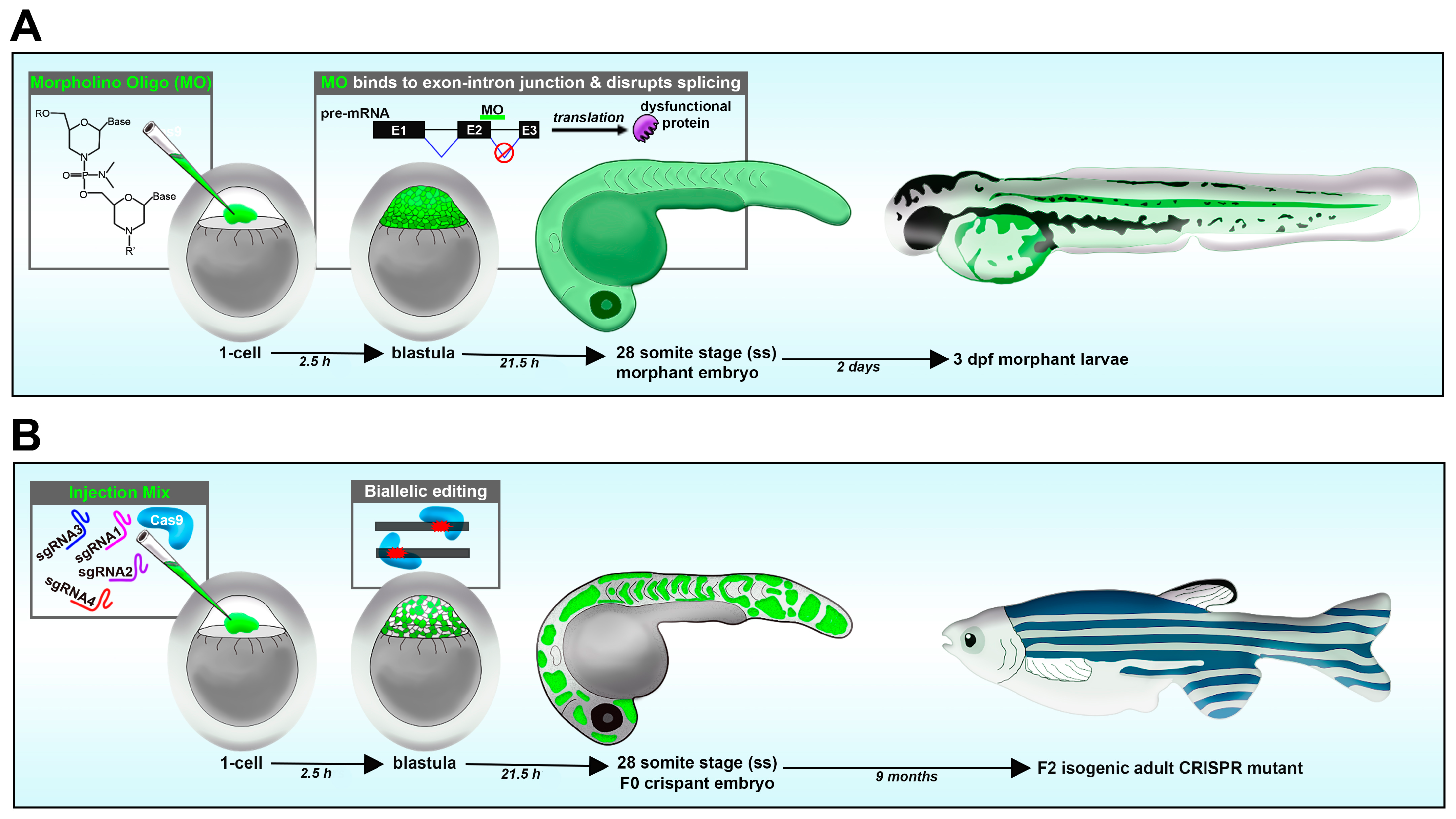
Figure 2. Reverse genetic loss-of-function strategies in the zebrafish embryo. (A) Morpholino knockdown. (B) CRISPR-Cas9.
References
- Preuss, H.G. Basics of renal anatomy and physiology. Clin. Lab. Med. 1993, 13, 1–11.
- Hoenig, M.P.; Zeidel, M.L. Homeostasis, the milieu intérieur, and the wisdom of the nephron. Clin. J. Am. Soc. Nephrol. 2014, 9, 1272–1281.
- Balzer, M.S.; Rohacs, T.; Susztak, K. How many cell types are in the kidney and what do they do? Annu. Rev. Physiol. 2022, 84, 507–531.
- Pollak, M.R.; Quaggin, S.E.; Hoenig, M.P.; Dworkin, L.D. The glomerulus: The sphere of influence. Clin. J. Am. Soc. Nephrol. 2014, 9, 1461–1469.
- Garg, P. A review of podocyte biology. Am. J. Nephrol. 2018, 47, 3–13.
- Zhuo, J.L.; Li, X.C. Proximal nephron. Compr. Physiol. 2013, 3, 1079–1123.
- Curthoys, N.P.; Moe, O.W. Proximal tubule function and response to acidosis. Clin. J. Am. Soc. Nephrol. 2014, 9, 1627–1638.
- Dantzler, W.H.; Layton, A.T.; Layton, H.E.; Pannabecker, T.L. Urine-concentrating mechanism in the inner medulla: Function of the thin limbs of the loops of Henle. Clin. J. Am. Soc. Nephrol. 2014, 9, 1781–1789.
- Mount, D.B. Thick ascending limb of the loop of Henle. Clin. J. Am. Soc. Nephrol. 2014, 9, 1974–1986.
- Subramanya, A.R.; Ellison, D.H. Distal convoluted tubule. Clin. J. Am. Soc. Nephrol. 2014, 9, 2147–2163.
- Pearce, D.; Soundararajan, R.; Trimpert, C.; Kashlan, O.B.; Deen, P.M.T.; Kohan, D.E. Collecting duct principal cell transport processes and their regulation. Clin. J. Am. Soc. Nephrol. 2015, 10, 135–146.
- Roy, A.; Al-bataineh, M.M.; Pastor-Soler, N.M. Collecting duct intercalated cell function and regulation. Clin. J. Am. Soc. Nephrol. 2015, 10, 305–324.
- Bertram, J.F.; Douglas-Denton, R.N.; Diouf, B.; Hughson, M.D.; Hoy, W.E. Human nephron number: Implications for health and disease. Pediatr. Nephrol. 2011, 26, 1529–1533.
- Luyckx, V.A.; Shukha, K.; Brenner, B.M. Low nephron number and its clinical consequences. Rambam Maimonides Med. J. 2011, 2, e0061.
- Black, M.J.; Sutherland, M.R.; Gubhaju, L.; Kent, A.L.; Dahlstrom, J.E.; Moore, L. When birth comes early: Effects on nephrogenesis. Nephrology (Carlton) 2013, 18, 180–182.
- McCampbell, K.K.; Wingert, R.A. Renal stem cells: Fact or science fiction? Biochem. J. 2012, 444, 153–168.
- Schedl, A. Renal abnormalities and their developmental origin. Nat. Rev. Genet. 2007, 8, 791–802.
- Dursun, H.; Bayazit, A.K.; Büyükçelik, M.; Soran, M.; Noyan, A.; Anarat, A. Associated anomalies in children with congenital solitary functioning kidney. Pediatr. Surg. Int. 2005, 21, 456–459.
- Westland, R.; Schreuder, M.F.; Ket, J.C.F.; van Wijk, J.A.E. Unilateral renal agenesis: A systematic review on associated anomalies and renal injury. Nephrol. Dial. Transplant. 2013, 28, 1844–1855.
- Vivante, A.; Kohl, S.; Hwang, D.; Dworschak, G.C.; Hildebrandt, F. Single-gene causes of congenital anomalies of the kidney and urinary tract (CAKUT) in humans. Pediatr. Nephrol. 2014, 29, 695–704.
- Rodriguez, M.M. Congenital anomalies of the kidney and the urinary tract (CAKUT). Fetal Pediatr. Pathol. 2014, 33, 293–320.
- Nicolaou, N.; Renkema, K.Y.; Bongers, E.M.; Giles, R.H.; Knoers, N.V. Genetic, environmental, and epigenetic factors involved in CAKUT. Nat. Rev. Nephrol. 2015, 11, 720–731.
- Romagnani, P.; Lasagni, L.; Remuzzi, G. Renal progenitors: An evolutionary conserved strategy for kidney regeneration. Nat. Rev. Nephrol. 2013, 9, 137–146.
- Peired, A.J.; Melica, M.E.; Molli, A.; Nardi, C.; Romagnani, P.; Lasagni, L. Molecular mechanisms of renal progenitor regulation: How many pieces in the puzzle? Cells 2021, 10, 59.
- Naved, B.A.; Bonventre, J.V.; Hubbell, J.A.; Hukriede, N.A.; Humphreys, B.D.; Kesselman, C.; Valerius, M.T.; McMahon, A.P.; Shankland, S.J.; Wertheim, J.A.; et al. Kidney repair and regeneration: Perspectives of the NIDDK (Re)Building a Kidney consortium. Kidney Int. 2022, 101, 845–853.
- Chambers, J.M.; Wingert, R.A. Advances in understanding vertebrate nephrogenesis. Tissue Barriers 2020, 8, e1832844.
- Little, M.H. Returning to kidney development to deliver synthetic kidneys. Dev. Biol. 2021, 474, 22–36.
- Beyenbach, K.W. Kidneys sans glomeruli. Am. J. Physiol. Renal Physiol. 2004, 286, F811–F827.
- Holz, P.H. Anatomy and physiology of the reptile renal system. Vet. Clin. N. Am. Exot. Anim. Pract. 2020, 23, 103–114.
- Orosz, S.E.; Echols, M.S. The urinary and osmoregulatory systems of birds. Vet. Clin. N. Am. Exot. Anim. Pract. 2020, 23, 1–19.
- Keogh, L.; Kilroy, D.; Bhattacharjee, S. The struggle to equilibrate outer and inner milieus: Renal evolution revisited. Ann. Anat. 2021, 233, 151610.
- Schnell, J.; Achieng, M.; Lindström, N.O. Principles of human and mouse nephron development. Nat. Rev. Nephrol. 2022, 18, 628–642.
- Senarat, S.; Kettratad, J.; Pairohakul, S.; Ampawong, S.; Huggins, B.P.; Coleman, M.M.; Kaneko, G. An update on the evolutionary origin of aglomerular kidney with structural and ultrastructural descriptions of the kidney in three fish species. J. Fish Biol. 2022, 100, 1283–1298.
- Evans, R.G. Evolution of the glomerulus in a marine environment and its implications for renal function in terrestrial vertebrates. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2023, 324, R143–R151.
- Gerlach, G.F.; Wingert, R.A. Kidney organogenesis in the zebrafish: Insights into vertebrate nephrogenesis and regeneration. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 559–585.
- Drummond, I.A.; Majumdar, A.; Hentschel, H.; Elger, M.; Solnica-Krezel, L.; Schier, A.F.; Neuhauss, S.C.; Stemple, D.L.; Zwartkruis, F.; Rangini, Z.; et al. Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development 1998, 125, 4655–4667.
- Majumdar, A.; Drummond, I.A. Podocyte differentiation in the absence of endothelial cells as revealed in the zebrafish avascular mutant, cloche. Dev. Genet. 1999, 24, 220–229.
- Majumdar, A.; Drummond, I.A. The zebrafish floating head mutant demonstrates podocytes play an important role in directing glomerular differentiation. Dev. Biol. 2000, 222, 147–157.
- Serluca, F.C.; Fishman, M.C. Pre-pattern in the pronephric kidney field of zebrafish. Development 2001, 128, 2233–2241.
- Hsu, H.; Lin, G.; Chung, B. Parallel early development of zebrafish interrenal glands and pronephros: Differential control by Wt1 and Ff1b. Development 2003, 130, 2107–2116.
- Drummond, B.E.; Ercanbrack, W.S.; Wingert, R.A. Modeling podocyte ontogeny and podocytopathies with the zebrafish. J. Dev. Biol. 2023, 11, 9.
- Kramer-Zucker, A.G.; Wiessner, S.; Jensen, A.M.; Drummond, I.A. Organization of the pronephric filtration apparatus in zebrafish requires Nephrin, Podocin and the FERM domain protein Mosaic eyes. Dev. Biol. 2005, 285, 316–329.
- Kramer-Zucker, A.G.; Olale, F.; Haycraft, C.J.; Yoder, B.K.; Schier, A.F.; Drummond, I.A. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer’s vesicle is required for normal organogenesis. Development 2005, 132, 1907–1921.
- Bollig, F.; Mehringer, R.; Perner, B.; Hartung, C.; Schäfer, M.; Schartl, M.; Volff, J.; Winkler, C.; Englert, C. Identification and comparative expression analysis of a second Wt1 gene in zebrafish. Dev. Dyn. 2006, 235, 554–561.
- Perner, B.; Englert, C.; Bollig, F. The Wilms tumor genes Wt1a and Wt1b control different steps during formation of the zebrafish pronephros. Dev. Biol. 2007, 309, 87–96.
- Ma, M.; Jiang, Y.J. Jagged2a-notch signaling mediates cell fate choice in the zebrafish pronephric duct. PLoS Genet. 2007, 3, e18.
- Liu, Y.; Pathak, N.; Kramer-Zucker, A.; Drummond, I.A. Notch signaling controls the differentiation of transporting epithelia and multiciliated cells in the zebrafish pronephros. Development 2007, 134, 1111–1122.
- Ikenaga, T.; Urban, J.M.; Gebhart, N.; Hatta, K.; Kawakami, K.; Ono, F. Formation of the spinal network in zebrafish determined by domain-specific pax genes. J. Comp. Neurol. 2011, 519, 1562–1579.
- Tena, J.J.; Neto, A.; de la Calle-Mustienes, E.; Bras-Pereira, C.; Casares, F.; Gómez-Skarmeta, J.L. Odd-Skipped genes encode repressors that control kidney development. Dev. Biol. 2007, 301, 518–531.
- Mudumana, S.P.; Hentschel, D.; Liu, Y.; Vasilyev, A.; Drummond, I.A. odd skipped related1 reveals a novel role for endoderm in regulating kidney versus vascular cell fate. Development 2008, 135, 3355–3367.
- Neto, A.; Mercader, N.; Gómez-Skarmeta, J.L. The Osr1 and Osr2 genes act in the pronephric anlage downstream of retinoic acid signaling and upstream of Wnt2b to maintain pectoral fin development. Development 2012, 139, 301–311.
- Tomar, R.; Mudumana, S.P.; Pathak, N.; Hukriede, N.A.; Drummond, I.A. Osr1 is required for podocyte development downstream of Wt1a. J. Am. Soc. Nephrol. 2014, 25, 2539–2545.
- Perens, E.A.; Garavito-Aguilar, Z.V.; Guio-Vega, G.P.; Peña, K.T.; Schindler, Y.L.; Yelon, D. Hand2 inhibits kidney specification while promoting vein formation within the posterior mesoderm. eLife 2016, 5, e19941.
- Perens, E.A.; Diaz, J.T.; Quesnel, A.; Crump, J.G.; Yelon, D. osr1 couples intermediate mesoderm cell fate with temporal dynamics of vessel progenitor cell differentiation. Development 2021, 148, dev198408.
- Drummond, B.E.; Chambers, B.E.; Wesselman, H.M.; Gibson, S.; Arceri, L.; Ulrich, M.N.; Gerlach, G.F.; Kroeger, P.T.; Leshchiner, I.; Goessling, W.; et al. osr1 maintains renal progenitors and regulates podocyte development by promoting wnt2ba via the antagonism of hand2. Biomedicines 2022, 10, 2868.
- Wingert, R.A.; Selleck, R.; Yu, J.; Song, H.; Chen, Z.; Song, A.; Zhou, Y.; Thisse, B.; Thisse, C.; McMahon, A.P.; et al. The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 2007, 3, e189.
- Ebarasi, L.; He, L.; Hultenby, K.; Takemoto, M.; Betsholtz, C.; Tryggvason, K.; Majumdar, A. A reverse genetic screen in the zebrafish identifies Crb2b as a regulator of the glomerular filtration barrier. Dev. Biol. 2009, 334, 1–9.
- Lyons, J.P.; Miller, R.K.; Zhou, X.; Weidinger, G.; Deroo, T.; Denayer, T.; Park, J.; Ji, H.; Hong, J.Y.; Li, A.; et al. Requirement of Wnt/Β-Catenin signaling in pronephric kidney development. Mech. Dev. 2009, 126, 142–159.
- de Groh, E.D.; Swanhart, L.M.; Cosentino, C.C.; Jackson, R.L.; Dai, W.; Kitchens, C.A.; Day, B.W.; Smithgall, T.E.; Hukriede, N.A. Inhibition of histone deacetylase expands the renal progenitor cell population. J. Am. Soc. Nephrol. 2010, 21, 794–802.
- Wingert, R.A.; Davidson, A.J. Zebrafish nephrogenesis involves dynamic spatiotemporal expression changes in renal progenitors and essential signals from retinoic acid and irx3b. Dev. Dyn. 2011, 240, 2011–2027.
- O’Brien, L.L.; Grimaldi, M.; Kostun, Z.; Wingert, R.A.; Selleck, R.; Davidson, A.J. Wt1a, Foxc1a, and the Notch Mediator Rbpj physically interact and regulate the formation of podocytes in zebrafish. Dev. Biol. 2011, 358, 318–330.
- Wang, H.; Lehtonen, S.; Chen, Y.C.; Heikkila, E.; Panula, P.; Holthofer, H. Neph3 associates with regulation of glomerular and neural development in zebrafish. Differentiation 2012, 83, 38–46.
- Ichimura, K.; Powell, R.; Nakamura, T.; Kurihara, H.; Sakai, T.; Obara, T. Podocalyxin regulates pronephric glomerular development in zebrafish. Physiol. Rep. 2013, 1, e00074.
- Naylor, R.W.; Przepiorski, A.; Ren, Q. HNF1B is essential for nephron segmentation during nephrogenesis. J. Am. Soc. Nephrol. 2013, 24, 77–87.
- Gerlach, G.F.; Wingert, R.A. Zebrafish pronephros tubulogenesis and epithelial identity maintenance are reliant on the polarity proteins Prkc iota and zeta. Dev. Biol. 2014, 396, 183–200.
- McKee, R.; Gerlach, G.F.; Jou, J.; Cheng, C.N.; Wingert, R.A. Temporal and spatial expression of tight junction genes during zebrafish pronephros development. Gene Expr. Patterns 2014, 16, 104–113.
- Fukuyo, Y.; Nakamura, T.; Bubenshchikova, E.; Powell, R.; Tsuji, T.; Janknecht, R.; Obara, T. Nephrin and Podocin functions are highly conserved between the zebrafish pronephros and mammalian metanephros. Mol. Med. Rep. 2014, 9, 457–465.
- Cheng, C.N.; Li, Y.; Marra, A.N.; Verdun, V.; Wingert, R.A. Flat mount preparation for observation and analysis of zebrafish embryo specimens stained by whole mount in situ hybridization. J. Vis. Exp. 2014, 89, 51604.
- Cheng, C.N.; Wingert, R.A. Nephron proximal tubule patterning and corpuscles of Stannius formation are regulated by the sim1a transcription factor and retinoic acid in zebrafish. Dev. Biol. 2015, 399, 100–116.
- Dong, L.; Pietsch, S.; Tan, Z.; Perner, B.; Sierig, R.; Kruspe, D.; Groth, M.; Witzgall, R.; Gröne, H.; Platzer, M.; et al. Integration of cistromic and transcriptomic analyses identifies Nphs2, Mafb, and Magi2 as Wilms’ Tumor 1 target genes in podocyte differentiation and maintenance. J. Am. Soc. Nephrol. 2015, 26, 2118–2128.
- Marra, A.N.; Wingert, R.A. Epithelial cell fate in the nephron tubule is mediated by the ETS transcription factors etv5a and etv4 during zebrafish kidney development. Dev. Biol. 2016, 411, 231–245.
- Zhu, X.; Chen, Z.; Zeng, C.; Wang, L.; Xu, F.; Hou, Q.; Liu, Z. Ultrastructural characterization of the pronephric glomerulus development in zebrafish. J. Morphol. 2016, 277, 1104–1112.
- Kroeger, P.T.; Drummond, B.E.; Miceli, R.; McKernan, M.; Gerlach, G.F.; Marra, A.N.; Fox, A.; McCampbell, K.K.; Leshchiner, I.; Rodriguez-Mari, A.; et al. The zebrafish kidney mutant zeppelin reveals that brca2/fancd1 is essential for pronephros development. Dev. Biol. 2017, 428, 148–163.
- Zhou, W.; Boucher, R.C.; Bollig, F.; Englert, C.; Hildebrandt, F. Characterization of mesonephric development and regeneration using transgenic zebrafish. Am. J. Physiol. Renal Physiol. 2010, 299, F1040–F1047.
- Diep, C.Q.; Peng, Z.; Ukah, T.K.; Kelly, P.M.; Daigle, R.V.; Davidson, A.J. Development of the zebrafish mesonephros. Genesis 2015, 53, 257–269.
- Reimschuessel, R. A fish model of renal regeneration and development. ILAR J. 2001, 42, 285–291.
- Diep, C.Q.; Ma, D.; Deo, R.C.; Holm, T.M.; Naylor, R.W.; Arora, N.; Wingert, R.A.; Bollig, F.; Djordjevic, G.; Lichman, B.; et al. Identification of adult nephron progenitors capable of kidney regeneration in zebrafish. Nature 2011, 470, 95–100.
- Davidson, A.J. Uncharted waters: Nephrogenesis and renal regeneration in fish and mammals. Pediatr. Nephrol. 2011, 26, 1435–1443.
- McCampbell, K.K.; Wingert, R.A. New tides: Using zebrafish to study renal regeneration. Transl. Res. 2012, 163, 109–122.
- McCampbell, K.K.; Springer, K.N.; Wingert, R.A. Analysis of nephron composition and function in the adult zebrafish kidney. J. Vis. Exp. 2014, 90, e51644.
- McCampbell, K.K.; Springer, K.N.; Wingert, R.A. Atlas of cellular dynamics during zebrafish adult kidney regeneration. Stem Cells Int. 2015, 2015, 547636.
- Kamei, C.N.; Gallegos, T.F.; Liu, Y.; Hukriede, N.; Drummond, I.A. Wnt signaling mediates new nephron formation during zebrafish kidney regeneration. Development 2019, 146, dev168294.
- Chen, J.; Yu, T.; He, X.; Fu, Y.; Dai, L.; Wang, B.; Wu, Y.; He, J.; Li, Y.; Zhang, F.; et al. Dual roles of hydrogen peroxide in promoting zebrafish renal repair and regeneration. Biochem. Biophys. Res. Commun. 2019, 516, 680–685.
- Gallegos, T.F.; Kamei, C.N.; Rohly, M.; Drummond, I.A. Fibroblast growth factor signaling mediates progenitor cell aggregation and nephron regeneration in the adult zebrafish kidney. Dev. Biol. 2019, 454, 44–51.
- Liu, X.; Yu, T.; Tan, X.; Jin, D.; Yang, W.; Zhang, J.; Dai, L.; He, Z.; Li, D.; Zhang, Y.; et al. Renal interstitial cells promote nephron regeneration by secreting prostaglandin E2. eLife 2023, 12, e81438.
- Lieschke, G.J.; Currie, P.D. Animal models of human disease: Zebrafish swim into view. Nat. Rev. Genet. 2007, 8, 353–367.
- Phillips, J.B.; Westerfield, M. Zebrafish models in translational research: Tipping the scales toward advancements in human health. Dis. Model Mech. 2014, 7, 739–743.
- Irion, U.; Nüsslein-Volhard, C. Developmental genetics with model organisms. Proc. Natl. Acad. Sci. USA 2022, 119, e2122148119.
- Howe, K.; Clark, M.; Torroja, C.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503.
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310.
- Lawson, N.D.; Wolfe, S.A. Forward and reverse genetic approaches for the analysis of vertebrate development in the zebrafish. Dev. Cell 2011, 21, 48–64.
- Molinari, E.; Sayer, J.A. Disease modeling to understand the pathomechanisms of human genetic kidney disorders. Clin. J. Am. Soc. Nephrol. 2020, 15, 855–872.
- Sun, Z.; Amsterdam, A.; Pazour, G.J.; Cole, D.G.; Miller, M.S.; Hopkins, N. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development 2004, 131, 4085–4093.
- Kroeger, P.T.; Poureetezadi, S.J.; McKee, R.; Jou, J.; Miceli, R.; Wingert, R.A. Production of haploid zebrafish embryos by in vitro fertilization. J. Vis. Exp. 2014, 89, 51708.
- Ebarasi, L.; Ashraf, S.; Bierzynska, A.; Gee, H.Y.; McCarthy, H.J.; Lovric, S.; Sadowski, C.E.; Pabst, W.; Vega-Warner, V.; Fang, H.; et al. Defects of CRB2 cause steroid-resistant nephrotic syndrome. Am. J. Hum. Genet. 2015, 96, 153–161.
- Gehrig, J.; Pandey, G.; Westhoff, J.H. Zebrafish as a model for drug screening in genetic kidney diseases. Front. Pediatr. 2018, 6, 183.
- Morales, E.E.; Wingert, R.A. Zebrafish as a model of kidney disease. Results Probl. Cell Differ. 2017, 60, 55–75.
- Skromne, I.; Prince, V.E. Current perspectives in zebrafish reverse genetics: Moving forward. Dev. Dyn. 2008, 237, 861–882.
- Nasevicius, A.; Ekker, S.C. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 2000, 26, 216–220.
- Draper, B.W.; Morcos, P.A.; Kimmel, C.B. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: A quantifiable method for gene knockdown. Genesis 2001, 30, 154–156.
- Doyon, Y.; McCammon, J.M.; Miller, J.C.; Faraji, F.; Ngo, C.; Katibah, G.E.; Amora, R.; Hocking, T.D.; Zhang, L.; Rebar, E.J.; et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 702–708.
- Meng, X.; Noyes, M.B.; Zhu, L.J.; Lawson, N.D.; Wolfe, S.A. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 695–701.
- Gupta, A.; Meng, X.; Zhu, L.J.; Lawson, N.D.; Wolfe, S.A. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011, 39, 381–392.
- Zhu, C.; Smith, T.; McNulty, J.; Rayla, A.L.; Lakshmanan, A.; Siekmann, A.F.; Buffardi, M.; Meng, X.; Shin, J.; Padmanabhan, A.; et al. Evaluation and application of modularly assembled zinc-finger nucleases in zebrafish. Development 2011, 138, 4555–4564.
- Miller, J.C.; Tan, S.; Qiao, G.; Barlow, K.A.; Wang, J.; Xia, D.F.; Meng, X.; Paschon, D.E.; Leung, E.; Hinkley, S.J.; et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011, 29, 143–148.
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55.
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405.
- Gupta, R.M.; Musunuru, K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J. Clin. Investig. 2014, 124, 4154–4161.
- Barrangou, R.; Marraffini, L.A. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol. Cell 2014, 54, 234–244.
- Jao, L.-E.; Wente, S.R.; Chen, W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909.
- Auer, T.O.; Duroure, K.; De Cian, A.; Concordet, J.-P.; Del Bene, F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014, 24, 142–153.
- Montague, T.G.; Cruz, J.M.; Gagnon, J.A.; Church, G.M.; Valen, E. CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014, 42, W401–W407.
- Tsai, S.Q.; Zheng, Z.; Nguyen, N.T.; Liebers, M.; Topkar, V.V.; Thapar, V.; Wyvekens, N.; Khayter, C.; Iafrate, A.J.; Le, L.P.; et al. GUIDE-Seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015, 33, 187–197.
- Hoffmann, S.; Mullins, L.; Rider, S.; Brown, C.; Buckley, C.B.; Assmus, A.; Li, Z.; Sierra Beltran, M.; Henderson, N.; Del Pozo, J.; et al. Comparative studies of renin-null zebrafish and mice provide new functional insights. Hypertension 2022, 79, e56–e66.
More
