Phosphate (Pi) transporters play critical roles in Pi acquisition and homeostasis. However, currently little is known about these genes in oil crops. In this study, we aimed to characterize the five Pi transporter gene families (PHT1-5) in allotetraploid
Brassica napus
. We identified and characterized 81 putative PHT genes in
B. napus
(
BnaPHTs
), including 45 genes in PHT1 family (
BnaPHT1s
), four
BnaPHT2s
, 10
BnaPHT3s
, 13
BnaPHT4s
and nine
BnaPHT5s
. Phylogenetic analyses showed that the largest PHT1 family could be divided into two groups (Group I and II), while PHT4 may be classified into five, Groups I-V. Gene structure analysis revealed that the exon-intron pattern was conservative within the same family or group. The sequence characteristics of these five families were quite different, which may contribute to their functional divergence. Transcription factor (TF) binding network analyses identified many potential TF binding sites in the promoter regions of candidates, implying their possible regulating patterns. Collinearity analysis demonstrated that most
BnaPHTs
were derived from an allopolyploidization event (~40.7%) between
Brassica rapa
and
Brassica oleracea
ancestors, and small-scale segmental duplication events (~39.5%) in the descendant. RNA-Seq analyses proved that many
BnaPHTs
were preferentially expressed in leaf and flower tissues. The expression profiles of most colinearity-pairs in
B. napus
are highly correlated, implying functional redundancy, while a few pairs may have undergone neo-functionalization or sub-functionalization during evolution. The expression levels of many
BnaPHTs
tend to be up-regulated by diifferent hormones inductions, especially for IAA, ABA and 6-BA treatments. qRT-PCR assay demonstrated that six
BnaPHT1s
(
BnaPHT1.11
,
BnaPHT1.14
,
BnaPHT1.20
,
BnaPHT1.35
,
BnaPHT1.41
,
BnaPHT1.44
) were significantly up-regulated under low- and/or rich- Pi conditions in
B. napus
roots.
- Brassica napusL.
- PHT gene family
- phylogenetic analysis
- expression analysis
1. Identification of PHT Genes in B. napusIdentification of PHT Genes in B. napus
To identify the
PHTs
in the
B. napus
genome, a preliminary BLASTP search was performed using the sequences of known
Arabidopsis
PHT proteins (AtPHTs) as queries, based on the available
B. napus
genome database in GENOSCOPE (Darmor–
bzh, http://www.genoscope.cns.fr/brassicanapus/) [1]. In each case, a large number of deduced PHT homologous sequences were acquired. The redundant sequences were firstly discarded from our dataset. Then the remaining sequences were verified by SMART (http://smart.embl-heidelberg.de/). The sequences possessing incomplete open reading frames (ORFs), especially long deletion were excluded from further analysis, such as BnaCnng51590D (Table S1). Finally, we obtained 81 typical PHT genes with relative complete ORFs in
, http://www.genoscope.cns.fr/brassicanapus/) [31]. In each case, a large number of deduced PHT homologous sequences were acquired. The redundant sequences were firstly discarded from our dataset. Then the remaining sequences were verified by SMART (http://smart.embl-heidelberg.de/). The sequences possessing incomplete open reading frames (ORFs), especially long deletion were excluded from further analysis, such as BnaCnng51590D (Table S1). Finally, we obtained 81 typical PHT genes with relative complete ORFs in
B. napus
(
BnaPHTs
), and temporarily named them according to their order on the corresponding chromosomes (Table S1). To our knowledge, the candidate
PHT
genes in
B. napus consist of the largest PHT family known to date [2][3][4][5]. This may attribute to
consist of the largest PHT family known to date [25–27,32]. This may attribute to
B. napus
(AACC. n=19) is an allotetraploid produced by the recent hybridization between
B. rapa
(AA.
n
=10) and
B. oleracea
(CC.
n
=9) about 7500 years ago.
Similarly, we also identified 46 candidate PHT homologs in
Brassica rapa
(
BrPHTs
) and 26 PHT homologs in
Brassica oleracea
(
BoPHTs
) from Phytozome v12 (https://phytozome.jgi.doe.gov/) by the same method (Table S2).
2. Phylogenetic Analysis of B. napus PHT Gene FamilyPhylogenetic Analysis of B. napus PHT Gene Family
To determine the evolutionary relationship of
B. napus
PHT gene families, we constructed a Neighbor-Joining (NJ) tree and a Maximum Likelihood (ML) trees of the candidate 165 PHT proteins from
B. napus
(80),
Arabidopsis
(22),
B. rapa
(42) and
B. oleracea
(21) based on the alignment of the full-length amino acid sequences using MEGA5.0 respectively. Five members (BnaPHT1.24, BrPHT1.25, BrPHT5.3, BoPHT1.4 and BoPHT1.5) were excluded from the phylogenetic trees because of technological reason (lacking common sites for the sequence pairs due to large sequence deletion).
The topologies and bootstrap support values of the NJ and ML trees were highly congruent (Figure 1 and Figure S1), and the candidate PHT members were clustered into five main clusters: PHT1-5 families (Figure 1). The PHT1 family is the largest one which includes nine genes in
Arabidopsis
(
AtPHT1s
), 25 in
B. rapa
(
BrPHT1s
), 8
B. oleracea
(
BoPHT1s
) and 44 in
B. napus
(
BnaPHT1s
); the PHT2 family contained one
AtPHT2
gene, two
BrPHT2s
, two
BoPHT2s
and four
BnaPHT2s
; the PHT3 family included three
AtPHT3s
, five
BrPHT3s
, four
BoPHT3s
and 10
BnaPHT3s
; the PHT4 family was consisted of six
AtPHT4s
, seven
BrPHT4s
, six
BoPHT4s
and 13
BnaPHT4s
; while the PHT5 family contained three
AtPHT5s
, three
BrPHT5s
, one
BoPHT5
gene and nine
BnaPHT5s (Figure 1). Consistent with previous studies [6], the PHT1 family could be further classified into two groups, where group I composed of 11
(Figure 1). Consistent with previous studies [33], the PHT1 family could be further classified into two groups, where group I composed of 11
BnaPHT1s
, seven
BrPHT1s
and five
BoPHT1s
that were homologous to
AtPHT1.8
and
AtPHT1.9
; group II contained 33
BnaPHT1s
, 18
BrPHT1s
and three
BoPHT1s
that were homologous to
AtPHT1.1
-
AtPHT1.7
. Similarly, the PHT4 family might be classified into five groups (Group I-Ⅴ) (Figure 1), where group I contained nine Brassica members and
AtPHT4.2
and
AtPHT4.3
; group II consisted of
AtPHT4.5
and Brassica homologs; group III included
AtPHT4.6
and four Brassica homologs; group IV composed of
AtPHT4.1
and four Brassica homologs
,
and group Ⅴ included
AtPHT4.4
and eight Brassica homologs.
Physicochemical property analysis showed that the molecular weight (MW) of candidate BnaPHT proteins (BnaPHTs) ranged from19.20-78.90 kDa, the isoelectric point (IP) is 5.44–10.01 (Table S1). The amino acid length of BnaPHTs varied from 279 to 704 aa, and these of PHT5 family are almost the longest while the PHT3 family are obviously shorter than the others. Subcellular localization analysis found that members of PHT1 and 4 families are located on the cell membrane, and members of PHT2 and 3 families are located on chloroplast and mitochondrion, respectively. In contrast, the majority of the PHT5 family are located on the cell membrane and vacuole except for one (
BnaPHT5.2), which was located on the cell membrane and chloroplast. The diverse subcellular localization features (Table S1) of these five families were highly consistent with the functional diversity of their homologs in plants [6].
, which was located on the cell membrane and chloroplast. The diverse subcellular localization features (Table S1) of these five families were highly consistent with the functional diversity of their homologs in plants [33].
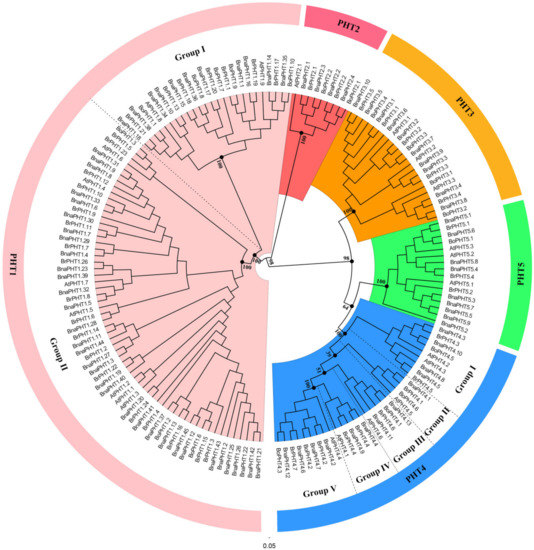
Figure 1.
Phylogenetic analysis of phosphate transporter (PHT) gene families in
Brassica napus
,
Arabidopsis
,
Brassica rapa
and
Brassica oleracea
. The colored background indicates different PHT family. The different groups within the PHT1 and 4 families were separated by dashed lines, respectively.
3. Gene Structure of PHT GenesGene Structure of PHT Genes
To gain insight into the structural feature of
PHTs
in
B. napus
,
Arabidopsis
,
B. rapa
and
B. oleracea
, we compared their full-length cDNA and genomic DNA sequences to determine the insertion site and number of introns within each gene. The results of
B. napus
and
Arabidopsis
PHTs
were viewed by GSDS software in Figure 2. The intron patterns of the five PHT families across these four species were presented in a schematic form in Figure S2, respectively.
As shown in Figure 2, the coding regions of most
PHTs
in
B. napus
and
Arabidopsis
are disrupted by introns varying from one to 14, except for 9 members in PHT1 family (Figure 2B). Among the five PHT families, the gene structures in PHT2, 3, and 5 families were highly conserved in terms of the exon-intron structure with only a few exceptions that may be attributed to low genome sequence quality, respectively (Figure 2B). The genes in PHT2 family possessed 2-3 introns, those in PHT3 family possessed 4-6 introns; and those in PHT5 family generally possessed 8-9 introns except for
BnaPHT5.2
,
BrPHT5.2
and
BrPHT5.3
(containing four or six introns). In contrast, the gene structures in PHT1 and 4 families were relatively less conserved. The
PHTs
in PHT1 family generally possessed 1-4 introns, where 21 members of the 54
PHT1s
have one intron, 14
PHT1s
contain two introns, ten
PHT1s
have three or four introns, respectively (Figure 2B). However, the gene structures were generally conserved in the same branch in PHT1 family that was consistent with their group classification in the phylogenetic tree, particularly the group I (Figure 2A), forming three conserved intron patterns across the four species investigated (Figure 2B). Similarly, the intron patterns of
PHTs
in PHT4 family could be summarized into -five conserved intron patterns (Figure 2B). Similar results were observed in
B. rapa
and
B. oleracea
PHTs
(Figure S2). These results indicated that the gain and loss of introns likely occurred in these two families during evolution.
Taken together, our results showed that the exon-intron patterns were generally conserved in each family or group in these five PHT families across the four species, indicating that each pattern shares a common ancestor.

Figure 2.
Gene structures of candidate PHT genes (
PHTs
) across different families. (
A
) Phylogenetic analysis of PHT families in
B. napus
and
Arabidopsis
. Colored background indicates genes belong to different families. The dashed lines represent the genes belong to different groups. (
B
) Gene structure of
PHTs
in
B. napus
(
BnaPHTs
) and
Arabidopsis
(
AtPHTs
). Exon was indicated by green boxes, transmembrane (TM) by red boxes, and the spaces between the colored boxes correspond to introns. Numbers 0, 1, 2 represent introns in phases 0, 1, and 2, respectively. The colored dots represent the conservative intron insertion patterns corresponding to the Figure S2, respectively.
4. Sequence Analysis of B. napus PHT ProteinsSequence Analysis of B. napus PHT Proteins
To analyze the sequence features of different PHT families in
B. napus
, we predicted the domains in the full-length protein sequences of the five PHT families using SMART software (Figure 3 and Table S3).
Our results showed that the protein structures of these five families were generally different. Among them, members of PHT1, 4 and 5 families display a similar domain architecture that they all contain the major facilitator superfamily (MFS-1, functions in transmembrane transport) and Sugar (and other) transporter (Sugar-tr, functions in transmembrane transport) domains. However, the sequence features of these two domains were not conservative among these three families with the MFS-1 and Sugar-tr domains are partial or even completely overlapped in these families. As shown in Figure 3, the MFS-1 domain (~400 aa) in PHT1 family is completely overlapped by the Sugar-tr domain (~480 aa) near the N-terminus. In contrast, the Sugar-tr domain is completely overlapped by the MFS-1 domain in PHT5 family (Figure 3). In addition, nearly all BnaPHT5s (except BnaPHT5.2) contained an additional highly conservative SYG1/Pho81/XPR1 (SPX, functions in phosphate transport) domain before the MFS-1 domain with a length of approximately 52 aa. In contrast, the MFS-1 and Sugar-tr domains are partial overlapped in PHT4 family. Furthermore, compared to PHT1 family, the MFS-1 domains were relatively highly conserved in both of PHT4 or PHT5 families, in contrast, the Sugar-tr domains were relatively poorly conserved in these two families. However, the sequence features of PHT2 and PHT3 were quite different from the PHT1, 4 and 5 families (Figure 3). The average lengths of the BnaPHT2s and BnaPHT3s were 584 aa and 337 aa, respectively. Moreover, the BnaPHT2s contained the highly conserved (PHO4, functions in inorganic phosphate transmembrane transport) domain near the C-terminus (approximately 400 aa in length), whereas the BnaPHT3s included two separated conservative Mitochondrial carrier (Mito-carr, substrate carrier proteins that are involved in energy transfer) domains at the N-terminus (about 90 aa for each domain).
The TMPRED software analyses showed that nearly all members of these five PHT families contained different numbers of transmembrane (TM) domains, suggesting this domain is important for their functions in plants. As shown in Figure 2B, all the BnaPHT1s contained 6-14 transmembrane (TM) domains; the BnaPHT2s contained 10-12 TM domains which are consistent with the previous report [7][8]; the BnaPHT4s and BnaPHT5s contained 8-14 and 11-13 TM domains, respectively; in addition, most of the BnaPHT3s possessed only 1-5 TM domains, but four members (BnaPHT3.3, BnaPHT3.4, BnaPHT3.8 and BnaPHT3.9) which belonged to the same branch with AtPHT3.2, and AtPHT3.2 lacked the TM domains. Because all the BnaPHT3s contained two mito-carr domains (including BnaPHT3.3, BnaPHT3.4, BnaPHT3.8 and BnaPHT3.9), the PHT3 family may rely on the mito-carr domain instead of the TM domain to function as Pi transporters. Furthermore, the sequence characteristics and spatial positions of the TM domains are generally conserved in each family or group. However, their locations were different across different families, such as those of BnaPHT5s were located at the C-terminal while those in BnaPHT1s were distributed in the full length ORF region.
The TMPRED software analyses showed that nearly all members of these five PHT families contained different numbers of transmembrane (TM) domains, suggesting this domain is important for their functions in plants. As shown in Figure 2B, all the BnaPHT1s contained 6-14 transmembrane (TM) domains; the BnaPHT2s contained 10-12 TM domains which are consistent with the previous report [5,20]; the BnaPHT4s and BnaPHT5s contained 8-14 and 11-13 TM domains, respectively; in addition, most of the BnaPHT3s possessed only 1-5 TM domains, but four members (BnaPHT3.3, BnaPHT3.4, BnaPHT3.8 and BnaPHT3.9) which belonged to the same branch with AtPHT3.2, and AtPHT3.2 lacked the TM domains. Because all the BnaPHT3s contained two mito-carr domains (including BnaPHT3.3, BnaPHT3.4, BnaPHT3.8 and BnaPHT3.9), the PHT3 family may rely on the mito-carr domain instead of the TM domain to function as Pi transporters. Furthermore, the sequence characteristics and spatial positions of the TM domains are generally conserved in each family or group. However, their locations were different across different families, such as those of BnaPHT5s were located at the C-terminal while those in BnaPHT1s were distributed in the full length ORF region.
Taken together, our results indicate that the sequence characteristics were conserved in each of the five PHT families or groups but were different across them, which may have contributed to functional divergence across different families.
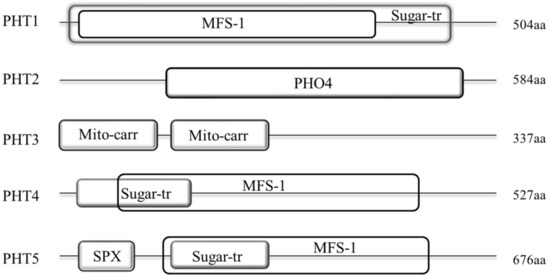
Figure 3.
Architecture of conserved protein domains in the five PHT families in
B. napus
. The conserved domains by Simple modular architecture research tool (SMART) were represented by different size boxes. The average protein length of each family is indicated on the right.
5. Chromosomal Distribution and Duplication of BnaPHTsChromosomal Distribution and Duplication of BnaPHTs
To investigate the gene duplication events within
B. napus
PHT families, we analyzed the chromosomal locations and collinearity relationship of candidates based on the annotion information in Genoscope database.
Our results showed that the candidate
BnaPHTs
were distributed on all the 19
B. napus
chromosomes, unevenly (Figure 4 and Table S1). The numbers of
BnaPHTs
between the two subgenomes are uneven, where the A
n
-subgenome has 40
BnaPHTs
while the C
n
-subgenome possess 34
BnaPHTs
. The genes distribution on each chromosome was bias, such as the A09 had a maximum number of nine genes, while A01 and A08 had a minimum of one gene. In addition, the chromosome information of seven genes was currently unknown (Table S1).
On the basis of the collinearity analyses between
B. napus
,
B. rape
and/or
B. oleracea
, we found that 65 of the 81
BnaPHTs
have colinear relationships with
BrPHTs
,
BoPHTs
and/or
BnaPHTs
(Table S5). Of these, ~50.8% of the colinear genes were inherited from
B. rapa
(22 genes, ~33.8%) or
B. oleracea
(11 genes, ~17.0%) genomes. Given that
B. napus
is a new allotetraploid that was hybridized by
B. rapa
and
B. oleracea
about 7500 years ago, these results demonstrated that most of the
BnaPHTs
(33/81, ~40.7%) were derived from its parents through allopolyploidy. Moreover, up to ~18.5%
BnaPHTs
were demonstrated to be derived from the segmental exchange (SE) event; ~7.4 % genes were from the homologous exchange (HE) event; and ~7.4% genes were from the segmental duplication (SD) event (Table S5), indicating this family tend to undergo small-scale duplication events in
B. napus
genome. Furthermore, we found that all the five PHT families have the largest number of genes that are orthologous pairs between
B. napus
and
B. rapa
or
B. oleracea
, including 14 pairs in PHT1 family, three pairs in PHT2 family, six pairs in PHT3 family, and five pairs in both of PHT4 and 5 families (Table S5). There are three, three and two pairs of
BnaPHT1s
involved in HE, SE, and SD events respectively; one pair of PHT2-4 families in HE event respectively; one pair of PHT3-5 families in SE event respectively; and one pair of PHT3 family, and three pairs of PHT4 family in SD event respectively (Table S5). In addition, five pairs of tandem duplication (TD) genes (
BnaPHT1.7/1.8
,
BnaPHT1.12/1.13
,
BnaPHT1.21/1.22
,
BnaPHT1.25/1.26
, and
BnaPHT1.30/1.31
) were identified (~6.2%), according to their distributions on chromosomes and sequence similarity (Table S5).
Overall, our results demonstrated that allopolyploid (large-scale duplication event) and small-scale duplication events after genomic heterogeneous doubling (HE, SE, and SD) were the main driving force for the large expansion of PHT gene family in
B. napus
genome. However, the trends of small-scale duplication events in the five PHT families were different.
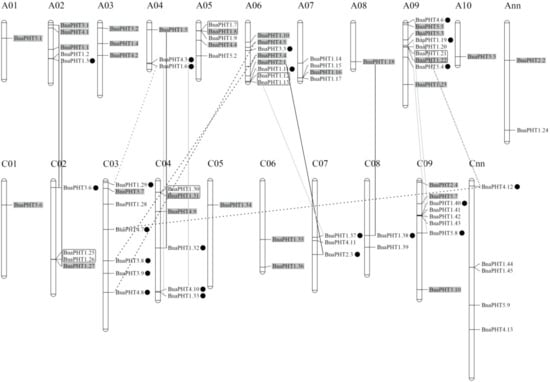
Figure 4.
Distribution of
PHT
genes on
B. napus
chromosomes. The 81
BnaPHTs
were mapped on the 19 chromosomes, except for seven genes. The Ann and Cnn chromosomes represent the chromosomal fragments mapped to A
n
or C
n
subgenome respectively, but the locations of these are as yet unclear. The genes in the gray background were originated from the whole genome duplication event (WGD). The black, gray and dashed lines represent the genes involved in homologous exchange (HE), segmental exchange (SE), and segmental duplication (SD) events with the new duplicated genes are marked in black dot, respectively. The genes in the black box represent tandem duplication (TD) pairs.
6. Transcription Factor Binding Network AnalysisTranscription Factor Binding Network Analysis
Gene expression and even function is commonly regulated by many kinds of transcription factor genes (TFs). To explore the transcriptional regulating mechanism of
BnaPHTs
, we analyzed the putative TFs binding sites in their promoter regions (upstream 1500 bp) by PlantTFDB database, and constructed their TF-binding network using Cytoscape software (Figure 5).
In general, various putative TF binding sites were observed in the promoter regions of candidate
BnaPHTs
, belonging to 30 TF gene families (Table S4). The most enriched TFs belong to the ERF (Ethylene responsive element binding factors, 63 genes), WRKY (WRKY DNA-binding protein, 19 genes), NAC (NAM, ATAF, and CUC TFs, 17 genes), MADS-box (MCM1, AG, DEF, and SRF TFs, 13 members), MYB (myeloblastosis, 15 genes, including 12 R2R3-MYB and three MYB-related genes) and Dof (DNA binding with one finger, 12 genes) families (Figure 5 A). Moreover, many TFs are likely to bind to the same target gene, such as the 63 candidate ERF TFs may target to 21
BnaPHTs
(Figure 5B). For example, many of the candidate WRKY TFs may bind to
BnaPHT1.11
and
BnaPHT1.44
genes promoters, suggesting these two
BnaPHTs
might be co-regulated by many WRKY TFs. Similarly,
BnaPHT1.42
and
BnaPHT4.7
were found to be the common targets of many ERF TFs, while many NAC TFs tend to target
BnaPHT1.15
and
BnaPHT5.4
(Figure 5B). In contrast, the Dof and MADS-box TFs are likely to target a much wider
BnaPHTs
, where the 13 MIKC-MADS-box TFs may regulate 27
BnaPHTs
and the 12 Dof TFs may regulate 28
BnaPHTs
(Figure 5B and Table S3). The remaining TF families only bind to a few
BnaPHTs
promoters as well, such as C3H TF may only bind to
BnaPHT5.5
promoter, EIL TF may bind to
BnaPHT1.41
promoter, while ZF-HD TF might bind to
BnaPHT1.16
promoter (Figure 5).
To date, several TF families were reported to be involved in the transcriptional regulation of
PHTs [9][10]. For example, the MYB-related gene
[34,35]. For example, the MYB-related gene
PHR1
(phosphate starvation response), regulated
PHT1
gene expression under low Pi condition through binding to the P1BS (PHR1-binding sequence) or P1BS-like domain; and the expressions of
AtPHT1;1
and
AtPHT1;4
genes were negatively regulated by
MYB62 [9]. Accordingly, our results showed that many MYB homologs may bind to
[34]. Accordingly, our results showed that many MYB homologs may bind to
BnaPHTs
promoters, suggesting their possible roles in Pi transport. It was previously reported that
WRKY45
could positively regulate
AtPHT1;1 expression under Pi starvation by binding to two W-boxes in its promoter [10]. Consequently, we found that 19 WRKY TFs may bind to
expression under Pi starvation by binding to two W-boxes in its promoter [35]. Consequently, we found that 19 WRKY TFs may bind to
BnaPHTs
promoters, implying possible regulating roles of this kind of TFs in
BnaPHTs
expression.
Our results indicate that the expressions of
BnaPHTs
may be regulated by many types of TFs, especially the ERF, WRKY, MYB, Dof and MADS-box family members.
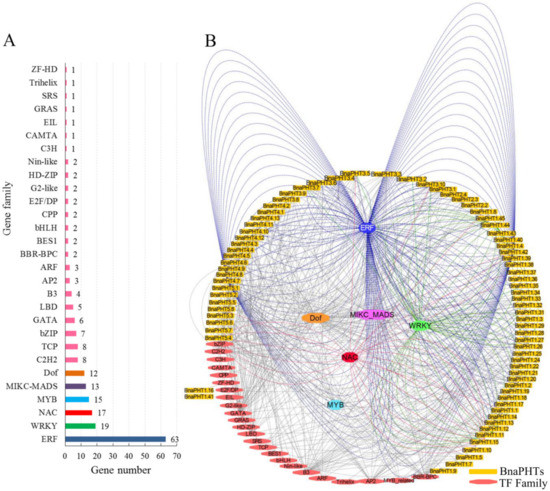
Figure 5.
Transcription factor (TF) binding network of
BnaPHTs
.
A
: The TF gene families that may bind to the promoter regions of
BnaPHTs
;
B
: The TFs binding network of
BnaPHTs
based on the anlayses in PlantTFDB database.
7. Spatial and Temporal Expressions of BnaPHTsSpatial and Temporal Expressions of BnaPHTs
Gene expression is related to its encoded protein function. So we inspected the expression patterns of the 81 candidate
BnaPHTs
in 50
B. napus
tissues/organs across different developmental stages, based on the RNA-seq data in NCBI (BioProject ID PRJNA358784). Twenty-seven
BnaPHTs
of PHT1 family with no or weak (FPKM < 1) expression levels that might be pseudogenes or have spatially or temporally restricted expressional pattern, were excluded from the heatmap.
In general, the majority of
BnaPHTs
were partial to highly express in leaf and flower tissues (Figure 6), indicating a temporal and a spatial expression trend. Consistent with their diverse sequence characteristics, the expression profiles of
BnaPHTs
in PHT1, 3 and 4 families are relative divergent, which may explain their possible functional division in phosphorus transport process. For instance, the expression pattern of PHT1 family could be classified into three main categories: the first type is preferentially expressed in germination seeds (
BnaPHT1.28
,
BnaPHT1.6
and
BnaPHT1.33
); the second type was highly expressed in radicle (
BnaPHT1.20
,
BnaPHT1.41
,
BnaPHT1.14
,
BnaPHT1.35
,
BnaPHT1.11
,
BnaPHT1.44, BnaPHT1.38
,
BnaPHT1.29
and
BnaPHT1.10
); while the remaining are mainly expressed in leaf and flower tissues. The expression profiles of
BnaPHT3s
can also be divided in to three categories: the first category was expressed in germination seeds and cotyledons, the second was mainly expressed in silique pericarps, and the rest was highly expressed in flower tissues. The expression profiles of the
BnaPHT3s
could be related to their predicted mitochondrial localization according to all of them contained the mito-carr domain. Similarly, the expressions of
BnaPHT4s
can be divided into two major groups: the first one consists of five members (
BnaPHT4.1, BnaPHT4.4, BnaPHT4.9, BnaPHT4.2
and
BnaPHT4.7
) that were highly expressed in leaf and flower tissues, while the second has relatively wider expression levels in the tissues investigated. In contrast, the expression profiles of the other two families are more conserved than the above three ones (Figure 6). In PHT2 family, the expressions of candidates are very similar and are highly expressed in leaf and flower tissues. Combined with their subcellular location analysis results, we predicted that they maybe affect Pi allocation at chloroplast. Similarly, the
BnaPHT5s
were mainly expressed in leaf, flower and silique pericarps tissues, and some members were also highly expressed in seed and embryo tissues. The expression patterns together with their subcellular location analysis suggested that the
BnaPHT5s
may function in Pi storage in many tissues.
In general, most of the
BnaPHTs
tend to be expressed in leaf and flower tissues. Moreover, the expression patterns within each family or group are similar, implying their functional conservation.
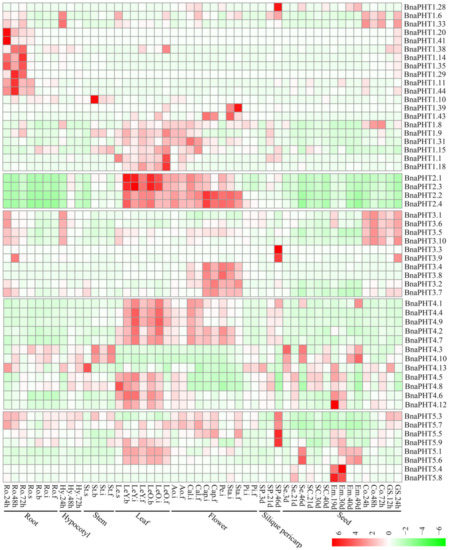
Figure 6.
Expression pattern of
BnaPHTs
in 50 tissues during plant development. Ro = root, St = stem, Le = leaf, Sp = silique pericarp, Sc = seed coat, Em = embryo, Ao = anthocaulus, Se = seed, Hy = hypocotyl, GS =germination seeds, Cap = capillament, Pi = pistil, Cal = calyx, Co = cotyledon, Pe = petal. The “h”, “d”, “i”, “f”, “s” indicate hour, day, seeding, budding, initial flowering, and full-bloom stages, respectively. The
BnaPHTs
with no or weak expression levels (FPKM < 1) were removed from the heatmap. The color bar at the low right represents log2 expression value (FPKM ≥ 1): green represents low expression, and red represents high expression.
8. Expression Analysis of BnaPHTs under Five Hormone InductionsExpression Analysis of BnaPHTs under Five Hormone Inductions
Plant hormones participate in the regulation of diverse plant processes. For instance, auxin is a key regulator for virtually every aspect of plant growth and development [11]; Gibberellin (GA) is a bioactive growth regulator which controls seed germination, stem elongation, flowering [12]; 6-Benzylaminopurine (6-BA) could stimulate cell division or cytokinesis [13]. Moreover, phytohormones are also known to be associated with nutrient signaling [14]. Therefore, we explored the expression patterns of
Plant hormones participate in the regulation of diverse plant processes. For instance, auxin is a key regulator for virtually every aspect of plant growth and development [36]; Gibberellin (GA) is a bioactive growth regulator which controls seed germination, stem elongation, flowering [37]; 6-Benzylaminopurine (6-BA) could stimulate cell division or cytokinesis [38]. Moreover, phytohormones are also known to be associated with nutrient signaling [39]. Therefore, we explored the expression patterns of
BnaPHTs
under five exogenous hormone treatments (IAA, ACC, ABA, GA
3
, and 6-BA) in
B. napus
roots, based on our RNA-Seq dataset (BioProject ID PRJNA608211). Forty-four
BnaPHTs
with no or weak expression levels (FPKM < 1) were excluded from the heatmap.
As mentioned above, the expressions of most
BnaPHTs
were generally low in roots (Figure 6). However, the expressions of many genes in roots were induced by the five hormone treatments (Figure 7). Among the five PHT families, the
BnaPHT1s
(e.g.,
BnaPHT1.1
,
BnaPHT1.6
,
BnaPHT1.9
, and
BnaPHT1.31
) were evidently up-regulated by ABA treatment; the
BnaPHT2s
(
BnaPHT2.1
-
BnaPHT2.4
) were up-regulated by all the five hormone treatments, especially IAA; the
BnaPHT3s
(
BnaPHT3.1
-
BnaPHT3.8
, and
BnaPHT3.10
) were up-regulated by IAA, ABA, GA
3
and/or 6-BA treatments; whereas the genes in PHT5 family (
BnaPHT5.1
,
BnaPHT5.3
,
BnaPHT5.5- BnaPHT5.7
, and
BnaPHT5.9
) were evidently up-regulated by IAA and/or ABA treatments (Figure 7). Morevoer, the expressions of homologs in a same family are generally similar, such as
BnaPHT1.1
,
BnaPHT1.9
and
BnaPHT1.31
having similar expression patterns under ABA treatment; and
BnaPHT2.1
-
BnaPHT2.4
having similar expression patterns under each of the five hormone inductions (Figure 7). These results suggest that the functions of
PHTs
in the same gene family are relative conserved. Notably, the expression patterns of
BnaPHT4s
were divided into two main patterns: the first type (
BnaPHT4.1
,
BnaPHT4.2
,
BnaPHT4.4
,
BnaPHT4.7
and
BnaPHT4.9
) was obviously up-regulated by IAA treatment, while another was up-regulated by IAA, ABA, GA
3
and/or 6-BA treatments under different trends. This suggests possibility functional divergence in the PHT4 family.
Previous reports have proven that the expressions of
PHTs by hormone-induction are similar to that by Pi stress, suggesting there is a linkage between hormone treatment and Pi starvation [15][16][17]. For instance, in rice,
by hormone-induction are similar to that by Pi stress, suggesting there is a linkage between hormone treatment and Pi starvation [40–42]. For instance, in rice,
OsPHT1;8 is involved in the cross-talk between Pi and auxin signaling, providing an evidence for the linkage between hormone and -Pi response [18]. Similarly, we found that the expressions of four
is involved in the cross-talk between Pi and auxin signaling, providing an evidence for the linkage between hormone and -Pi response [43]. Similarly, we found that the expressions of four
BnaPHT1s
(
BnaPHT1.6
,
BnaPHT1.9
,
BnaPHT1.31
and
BnaPHT1.33) which were up-regulated under Pi deficiency [19], were also up-regulated by IAA treatment (Figure 7). This implies a linkage between IAA treatment and Pi starvation. Moreover, cytokinin has been shown to be involved in the suppression of Pi starvation-induced gene expression [20]. Consistently, our results showed that the expressions of most
) which were up-regulated under Pi deficiency [29], were also up-regulated by IAA treatment (Figure 7). This implies a linkage between IAA treatment and Pi starvation. Moreover, cytokinin has been shown to be involved in the suppression of Pi starvation-induced gene expression [44]. Consistently, our results showed that the expressions of most
BnaPHT1s
were suppressed by 6-BA treatment (Figure 7), indicating similar mechanism may exist in
B. napus
.
In summary, our results demonstrate that exogenous hormones could strongly induce the expressions of many
BnaPHTs
in
B. napus
roots (especially IAA and ABA), indicating that hormones have an important role in their actions. Further research is required to elucidate their precise roles in hormone and −Pi signaling.
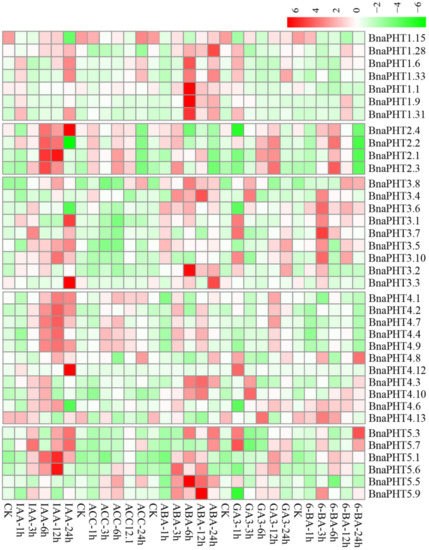
Figure 7
. Expression profiles of
BnaPHTs
under five hormone inductions in
B. napus
seedling roots by RNA-seq. CK: no extra hormone inductions (0 h), IAA: indoleacetic acid, ACC: 1-amino cyclopropanecarboxylic acid, ABA: abscisic acid, GA
3
: gibberellin acid 3, 6-BA: cytokinin. The “1 h”, “3 h”, “6 h”, “12 h”, and “24 h” represent hours after treatment. The color bar in the upper right represents log2 expression values (FPKM ≥ 1): green represents low expression and red represents high expression.
9. Expression Analysis of BnaPHTs in Response to High-Pi and Low-Pi Conditions by qRT-PCRExpression Analysis of BnaPHTs in Response to High-Pi and Low-Pi Conditions by qRT-PCR
The transcription levels of
PHTs are generally responsive to Pi levels. Among the five PHT families, members of PHT1 family are high-affinity Pi transporters in plants [2][21][22][23] and are up-regulated under Pi deprivation conditions [24][25]. On the basis of our spatiotemporal expression analysis (Figure 6) and the known functions of
are generally responsive to Pi levels. Among the five PHT families, members of PHT1 family are high-affinity Pi transporters in plants [13,14,18,45] and are up-regulated under Pi deprivation conditions [2,46]. On the basis of our spatiotemporal expression analysis (Figure 6) and the known functions of
Arabidopsis
homologs, three homologous pairs (
BnaPHT1.20
/
BnaPHT1.41
,
BnaPHT1.11
/
BnaPHT1.44
and
BnaPHT1.14
/
BnaPHT1.35
) which were highly expressed in roots were selected for further qRT-PCR assay to explore their potential roles in response to low Pi (1 μM, −Pi) and high Pi (2 mM, +Pi) conditions in
B. napus
seedling roots.
As shown in Figure 8, all of the six genes were strongly up-regulated under −Pi condition, but were relatively less induced by +Pi stress (except for
BnaPHT1.41
), indicating they were more sensitive to −Pi condition. The expression patterns of these genes can be classified into three patterns:
BnaPHT1.41
and
BnaPHT1.11
were strongly up-regulated under both of −Pi and +Pi conditions; the two homologs of
AtPHT1.9
,
BnaPHT1.14
and
BnaPHT1.35
were up-regulated by -Pi stress while were slightly up-regulated under +Pi condition; and
BnaPHT1.20
and
BnaPHT1.44
were strongly up-regulated under -Pi condition but were not induced by +Pi stress. Notably, as a sister pair, the expression profiles of
BnaPHT1.11
and
BnaPHT1.44
under −Pi and +Pi conditions were somewhat different, where the expression of
BnaPHT1.11
was gradually increased under −Pi treatment (except for on the fifth day), while
BnaPHT1.44
had the highest expression level on the third day under −Pi condition (Figure 8). This indicates functional divergence trend of sister pair genes during evolution.
Overall, our results demonstrated that the six
BnaPHTs
in PHT1 family may involve in −Pi stress response in
B. napus
seedling roots, which provided a fundament for further gene functional research.
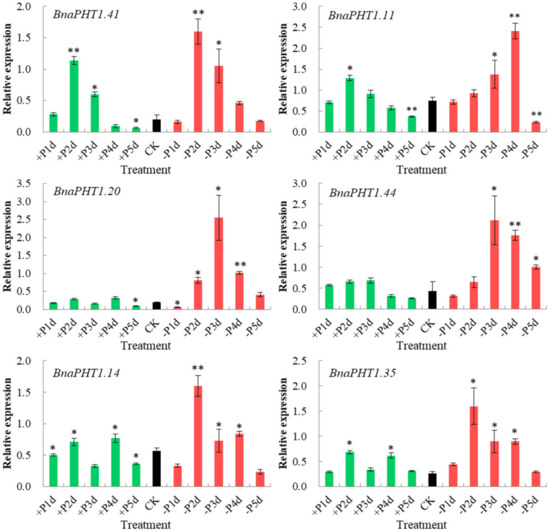
Figure 8.
Expressions of six
BnaPHT1s
under low- or high- Pi treatments by qRT-PCR. The transcript levels were determined in
B. napus
seedling roots by qRT-PCR method under low (-Pi) or high Pi (+Pi) conditions. CK: normal Pi condition; +P1d~+P5d: +Pi condition on 1-5 day (s); -P1d~-P5d: -Pi condition on 1~5 day (s). The
B. napus
Actin7
(
BnActin7
) (GenBank accession no. AF024716) was used as the reference gene. The red bar represents the expressions of
BnaPHTs
under -Pi conditions; the green bar represents the expressions of
BnaPHTs
under +Pi conditions; and the black bar represents the expressions of
BnaPHTs under normal Pi conditions (CK). Error bars indicate the standard deviation of three independent experiments. *: Significant difference (0.05 >
under normal Pi conditions (CK). Error bars indicate the standard deviation of three independent experiments. *: Significant difference (0.05 > p > 0.01); **: Extremely significant difference (p < 0.01).
25. Okumura, S.; Mitsukawa, N.; Shirano, Y.; Shibata, Phosphate transporter gene family of Arabidopsis thaliana. DNA Res. 1998, 5, 261–269, doi:10.1093/dnares/5.5.261.
26. Liu, F.; Chang, X.J.; Ye, Y.; Xie, W.B.; Wu, P.; Lian, X.M. Comprehensive sequence and whole-life-cycle expression profile analysis of the phosphate transporter gene family in rice. Plant 2011, 4, 1105–1122, doi:10.1093/mp/ssr058.
27. Zhang, C.; Meng, S.; Li, M.; Zhao, Genomic identification and expression analysis of the phosphate transporter gene family in poplar. Front. Plant Sci. 2016, 7, 1398, doi:10.3389/fpls.2016.01398.
28. Ding, G.D.; Zhao, Z.K.; Liao, Y.; Hu, Y.F.; Shi, L.; Long, Y. Quantitative trait loci for seed yield and yield related traits and their responses to reduced phosphorus supply in Brassica napus. Bot. 2012, 109, 747–759, doi:10.1093/aob/mcr323.
29. Li, Y.; Wang, X.; Zhang, H.; Wang, S.; Ye, X.; Shi, L.; Xu, F.; Ding, G. Molecular identification of the phosphate transporter family 1 (PHT1) genes and their expression profiles in response to phosphorus deprivation and other abiotic stresses in Brassica napus. PLoS ONE 2019, 14, e0220374, doi:10.1371/journal.pone.0220374.
30. Ren, F.; Zhao, C.Z.; Liu, C.S.; Huang, K.L.; Guo, Q.Q.; Chang, L.L.; Xiong, H.; Li, X.B. A Brassica napus PHT1 phosphate transporter, BnaPHT1;4, promotes phosphate uptake and affects roots architecture of transgenic Arabidopsis. Plant Mol. Biol. 2014, 86, 595, doi:10.1007/s11103-014-0249-y.
31. Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953, doi:10.1126/science.1253435.
32. Sun, T.; Li, M.; Shao, Y.; Yu, L.; Ma, F. Comprehensive Genomic Identification and Expression Analysis of the Phosphate Transporter (PHT) Gene Family in Apple. Plant Sci. 2017, 8, 426, doi:10.3389/fpls.2017.00426.
33. Wang, D.; Lv, S.; Jiang, P.; Li, Roles, regulation, and agricultural application of plant phosphate transporters. Front. Plant Sci. 2017, 8, 817, doi:10.3389/fpls.2017.00817.
34. Devaiah, B.N.; Madhuvanthi, R.; Karthikeyan, A.S.; Raghothama, K. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol. Plant 2009, 2, 43–58, doi:10.1093/mp/ssn081.
35. Wang, H.; Xu, Q.; Kong, Y.H.; Chen, Y.; Duan, J.Y.; Wu, W.H. Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER11 expression in response to phosphate starvation. Plant Physiol. 2014, 164, 2020–2029, doi:10.1104/pp.113.235077.
36. Ljung, K. Auxin metabolism and homeostasis during plant development. Development. 2013, 140, 943–950, doi:10.1242/dev.086363.
37. Yamaguchi, S. Gibberellin metabolism and its regulation. Rev. Plant Biol. 2008, 59, 225–251, doi:10.1146/annurev.arplant.59.032607.092804.
38. Deng, Y.; Zhao, L.; Zhang, L. Effects of sand and hormone treatment on seed germination of Bupleurum Chinese DC. And B. Falcatum, L. Zhongguo Zhong Yao Za Zhi. 1996, 21, 208–255.
39. Baek, D.; Chun, H.J.; Yun, D.J.; Kim, M.C. Cross-talk between phosphate starvation and other environmental stress signaling pathways in plants. Cells 2017, 40, 697–705, doi:10.14348/molcells.
40. Bates, T.R.; Lynch, J.P. Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Cell Environ. 1996, 19, 529–538, doi:10.1111/j.1365-3040.1996.tb00386.x.
41. Lynch, J.; Brown, K.M. Ethylene and plant responses to nutritional stress. Physiol. Plant 1997, 100, 613–619.
42. Gilbert, G.A.; Knight, J.D.; Vance, C.P.; Allan, D.L. Proteoid root development of phosphorus deficient lupin is mimicked by auxin and phosphonate. Ann. Bot. 2000, 85, 921–928.
43. Jia, H.; Zhang, S.; Wang, L.; Yang, Y.; Zhang, H.; Cui, H.; Shao, H.; Xu, G. OsPht1;8, a phosphate transporter, is involved in auxin and phosphate starvation response in rice. Exp.
> 0.01); **: Extremely significant difference (
Bot. 2017, 68, 5057–5068, doi:10.1093/jxb/erx317.
44. Martin, A.C.; del Pozo, J.C.; Iglesias, J.; Rubio, V.; Solano, R.; de la Pena, A.; Leyva, A.; Paz-Ares, J. Influence of cytokininson the expression of phosphate starvation responsive genes in Plant J. 2000, 24, 559–567, doi:10.1046/j.1365-313x.2000.00893.x.
45. Remy, E.; Cabrito, T.R.; Batista, R.A.; Teixeira, M.C.; Sá-Correia, I.; Duque, P. The Pht19 and Pht18 transporters mediate inorganic phosphate acquisition by the Arabidopsis
< 0.01).
thaliana root during phosphorus starvation. New Phytol. 2012, 195, 356–371, doi:10.1111/j.1469-8137.2012.04167.x.
46. Smith, S.E.; Jakobsen, I.; Grønlund, M.; Smith, F.A. Roles of arbuscular mycorrhizas in plant phosphorus nutrition: Interactions between pathways of phosphorus uptake in arbuscular mycorrhizal roots have important implications for understanding and manipulating plant phosphorus acquisition. Plant Physiol. 2011, 156, 1050–1057, doi:10.1104/pp.111.174581.
