Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Dean Liu and Version 1 by PAVAN KUMAR.
Hybridization is the most widely used modification technique for the dimension reduction problem. There are three types of hybridization: integrating a nature-inspired algorithm with another nature-inspired algorithm, integrating a nature-inspired algorithm with a classifier, and integrating a nature-inspired algorithm with filter or extraction techniques.
- nature-inspired algorithm
- high-dimensional data
- dimension reduction
1. Introduction
Nowadays, to address the issue of dimension reduction, many researchers use a variety of hybrid approaches with various combinations to speed up computation and gain the advantages of the various dimension reduction techniques [1]. Researchers used various combinations of algorithms in accordance with the demands of various data sets by integrating various feature selection and extraction techniques. Utilizing the most effective techniques is essential because resources and time are both finite. Nature-inspired metaheuristic algorithms have recently gained popularity as a tool for discovering significant features from large and complex biomedical datasets, as compared to other algorithms. Nature-inspired techniques include a wide range, from local learning techniques to global search solutions. Exploration and exploitation are the two key working processes in every nature-inspired metaheuristic algorithm. The exploitation process is looking for the best option from the currently available options, while exploration is discovering a novel solution on a large scale. Nature-inspired algorithms are founded on the diversification principle and take some clues from the environment to solve problems. Several nature-inspired techniques produce acceptable results in solving the problems of different domains [2]. Nature-inspired algorithms provide better models with fast convergence rates, quicker response times, fair exploration and exploitation, and fewer algorithm-specific control parameters.
There have been various reviews that cover numerous dimension reduction strategies for high-dimensional biomedical data. Most of the previously published surveys focused only on feature selection and extraction algorithms for DNA microarray datasets [3][4][5]. To the best of our knowledge, none of them provide a comprehensive data reduction algorithm, including feature selection, feature extraction, and their hybrid from the standpoint of ML algorithms used. The present review bridges the gap identified above, investigating the severe difficulty of dealing with high-dimensional gene expression datasets and several proposed solutions with nature-inspired algorithms. A taxonomy of data reduction techniques with the two most important processes of data reduction: feature selection and feature extraction, and their hybrid algorithms with nature-inspired techniques, have been defined. Therein, each class is further classified as supervised, unsupervised, or based on the applied machine-learning algorithm.
The human body is comprised of thousands of genes, although only a few are associated with a cancer diagnosis or complications in the human body. Therefore, for identifying the associated genes in a typical cancer type from any high-dimensional gene expression data, precisely, 2n possible subsets of genes are available for a dataset comprising n features [6][7]. Therefore, a successful dimensionality reduction algorithm provided the minimum number of genes for the learning techniques. A learning approach tends to generalize when it has numerous features, which causes its performance to degrade. There are two methods, that are regularly used to reduce dimensionality: first feature selection and second feature extraction. In order to attain improved learning performance, we required lower computational expenses and significantly improved interpretability. Dimension reduction attempts to choose just a few useful genes from the original set in accordance with such relevance evaluation criteria. Traditionally, the adoption of categorization characteristics seeks to pick the smallest collection of properties that should satisfy categorization accuracy [8]. Figure 31 shows the benefits of dimension reduction methods.
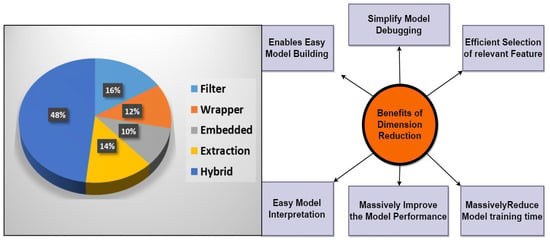
Figure 31.
Benefits of the dimension reduction method.
2. Feature (Gene) Selection Methods
Feature selection approaches have been split into three classes based on the selection technique: filter, wrapper, and embedded. Without employing any learning models, the feature selection picks the features depending on the data’s statistical properties.
2.1. Filter
Filter methods are used to determine the significance of any feature and fall under the univariate and multivariate categories. The univariate filter method first employs a specific criterion to pinpoint the most significant k-feature rankings, after which each attribute is evaluated and given a distinct rating. This type of filter, in contrast to a multivariate method, is unable to spot duplicate features since it ignores feature dependencies that affect classification results. Univariate filters, on the other hand, are quicker than multivariate ones. The most well-known univariate filters employed in several of the evaluations on this list were Fisher Score, Mutual Information, and Laplacian Score (LS). The multivariate filter takes into consideration the relationships between various qualities. As a result, this type of filter has superior classification performance than other types of filters and can evaluate both redundant and irrelevant material. Some of the survey studies utilized the most well-known univariate filters, Relief-f, minimal redundancy, and maximum relevance [9][10].
2.2. Wrapper
This feature selection approach relies on classifiers. It operates by choosing a subset of characteristics from a given learning model that yields the best outcomes. Wrappers fall within the stochastic and greedy search strategy categories. A greedy algorithm follows the problem-solving process by making the local optimum choice at each stage. On the other hand, the stochastic search technique is based on the concept of various unpredictable, nature-inspired methods and uses a variety of meta-heuristic algorithms.
2.3. Embedded
In an embedded method, feature selection is constructed into the classifier algorithm. During the training step, the classifier adjusts its internal parameters and determines the appropriate feature that produces the best classification accuracy.
3. Feature Extraction
Feature extraction is an intelligent substitute for feature selection to reduce the size of high-dimensional data. In the literature, it is also called feature construction or projection onto a low-dimensional subspace. The feature extraction method transforms the original feature into a lower-dimensional space; in this way, the problem is represented in a more discriminating (informative) space that makes further analysis more efficient. There are two main types of feature extraction algorithms: linear and nonlinear. Linear methods are usually faster, more robust, and more interpretable than non-linear methods. On the other hand, non-linear methods can sometimes discover the complicated structures of data where linear methods fail to distinguish [11].
4. Hybrid
The hybrid method of dimension reduction combines the benefits of feature extraction and feature selection approaches to improve the classification and prediction accuracy of an ML-based model. This kind of feature selection technique is often used in two phases. Filter techniques, whether a combination of filters or extraction, were used in the first stage [6][7][12][13] to limit the number of features conveyed to the wrapper stage, where classification performance will be improved with a suitable calculation time. They consequently inherit the remarkable computational speed of wrapper algorithms along with the computational effectiveness of filter algorithms. The first stage filter or extraction method has frequently been used to remove redundant genes in the hybrid method with two stages. In the second stage, wrapper algorithms take into account the learning algorithm to choose the biomarker gene subspace from among previously chosen features. The chosen genes were then used to create a prediction model, which was then tested for effectiveness. There have been numerous hybrid-based supervised feature selection methods studied. Hybrid algorithms, which combine filter or extraction with wrapper methods, have been developed in various studies [14][15][16]. Table 1 depicts the advantages and drawbacks of different types of dimension reduction techniques.
Table 1. Comparison of different dimension reduction techniques.
| Study Types | Advantages | Drawbacks | Regular Search | Effectiveness |
|---|---|---|---|---|
| Filter Method | Autonomous Neglects classification Quick calculation times |
No communication with the classification model Disregards important features |
Univariate Multivariate |
Quicker than other feature selection techniques lowers the feature’s significance |
| Wrapper Method | Dependent on feature Computerized and time consuming Interacts with the classifier and feature selection |
Overfitting risk Dependent on the classifier Lengthy calculation time Time that is complex exponential overfitting risk Dependent on the classifiers |
Deterministic Stochastic |
Superior to the filter method Extraordinary efficiency |
| Embedded Method | Having a low chance of overfitting Interacting with the classifier Using the best FS method with the classification model |
Classifier depends on the technique of selection Propensity for overfitting |
Integrated model Simplified model |
Computations are less expensive than wrapping |
| Feature Extraction | Higher discriminating power Control over fitting problem |
Loss of data interpretability The transformation may be expensive |
PCA, Linear discriminant analysis, ICA |
Used in an effective way in the hybrid algorithm |
| Hybrid Method | Combines multiple feature selection and extraction techniques. |
Time-consuming and challenging | Searching in depth Ideal FS |
Complexity Reduced mistake |
References
- Musheer, R.A.; Verma, C.K.; Srivastava, N. Novel machine learning approach for classification of high-dimensional microarray data. Soft Comput. 2019, 23, 13409–13421.
- Ramachandran, A.; Rustum, R.; Adeloye, A.J. Review of anaerobic digestion modeling and optimization using nature-inspired techniques. Processes 2019, 12, 953.
- Mohamad, A.B.; Zain, A.M.; Nazira Bazin, N.E. Cuckoo search algorithm for optimization problems a literature review and its applications. Appl. Artif. Intell. 2014, 28, 419–448.
- Aziz, R.; Verma, C.K.; Srivastava, N. Artificial neural network classification of high dimensional data with novel optimization approach of dimension reduction. Ann. Data Sci. 2018, 5, 615–635.
- Naik, M.; Nath, M.R.; Wunnava, A.; Sahany, S.; Panda, R. A new adaptive Cuckoo search algorithm. In Proceedings of the 2015 IEEE 2nd International Conference on Recent Trends in Information Systems (ReTIS), Kolkata, India, 9–11 July 2015; IEEE: New York, NY, USA; pp. 1–5.
- Aziz, R.; Verma, C.K.; Jha, M.; Srivastava, N. Artificial neural network classification of microarray data using new hybrid gene selection method. Int. J. Data Min. Bioinform. 2017, 17, 42–65.
- Tufail, A.B.; Ma, Y.K.; Kaabar, M.K.; Martínez, F.; Junejo, A.R.; Ullah, I.; Khan, R. Deep learning in cancer diagnosis and prognosis prediction: A minireview on challenges, recent trends, and future directions. Comput. Math. Methods Med. 2021, 2021, 9025470.
- Elemento, O.; Leslie, C.; Lundin, J.; Tourassi, G. Artificial intelligence in cancer research, diagnosis and therapy. Nat. Rev. Cancer 2021, 21, 747–752.
- Aziz, R.; Srivastava, N.; Verma, C.K. T-independent component analysis for svm classification of DNA-microarray data. Int. J. Bioinform. Res. 2015, 6, 305–312.
- Desai, N.P.; Baluch, M.F.; Makrariya, A.; Musheer Aziz, R. Image processing model with deep learning approach for fish species classification. Turk. J. Comput. Math. Educ. 2022, 13, 85–99.
- Alomari, O.A.; Khader, A.T.; Al-Betar, M.A.; Abualigah, L.M. Gene selection for cancer classification by combining minimum redundancy maximum relevancy and bat-inspired algorithm. Int. J. Data Min. Bioinform. 2017, 19, 32–51.
- Diao, R.; Shen, Q. Nature inspired feature selection meta-heuristics. Artif. Intell. Rev. 2015, 44, 311–340.
- Aziz, R.M. Cuckoo search-based optimization for cancer classification: A new hybrid approach. J. Comput. Biol. 2022, 29, 565–584.
- Aziz, R.M.; Hussain, A.; Sharma, P.; Kumar, P. Machine learning based soft computing regression analysis approach for crime data prediction. Karbala Int. J. Mod. Sci. 2022, 8, 1–19.
- Aziz, R.M.; Baluch, M.F.; Patel, S.; Kumar, P. A based approach to detect the Ethereum fraud transactions with limited attributes. Karbala Int. J. Mod. Sci. 2022, 8, 139–151.
- Luan, J.; Yao, Z.; Zhao, F.; Song, X. A novel method to solve supplier selection problem: Hybrid algorithm of genetic algorithm and ant colony optimization. Math. Comput. Simul. 2019, 156, 294–309.
More
