The balance of proteins in cells, is proteostasis (protein homeostasis). While protein balances can be moderated by cell division, in non-dividing cells, with age there is declining proteostasis and the accumulation of deleterious proteins can present as major problems, such as Alzheimer's disease, Parkinson's disease, Huntington's disease, etc. Budding yeast also exhibit ageing and can offer convenient means to discover more about biological pathways involved in proteostasis. Similarities of yeast and humans mean that yeast can provide valuable insights into multiple pathways of proteostasis, and can be used as a model for analysing drugs that may improve proteostasis and counter age-related decline in neuronal health.
- Alzheimer's disease
- amyloid beta
- yeast models
- homeostasis
- ageing
- proteostasis
- Saccharomyces cerevisiae
- neurodegeneration
- protein turnover
- toxic proteins
Note:All the information in this draft can be edited by authors. And the entry will be online only after authors edit and submit it.
1. Introduction
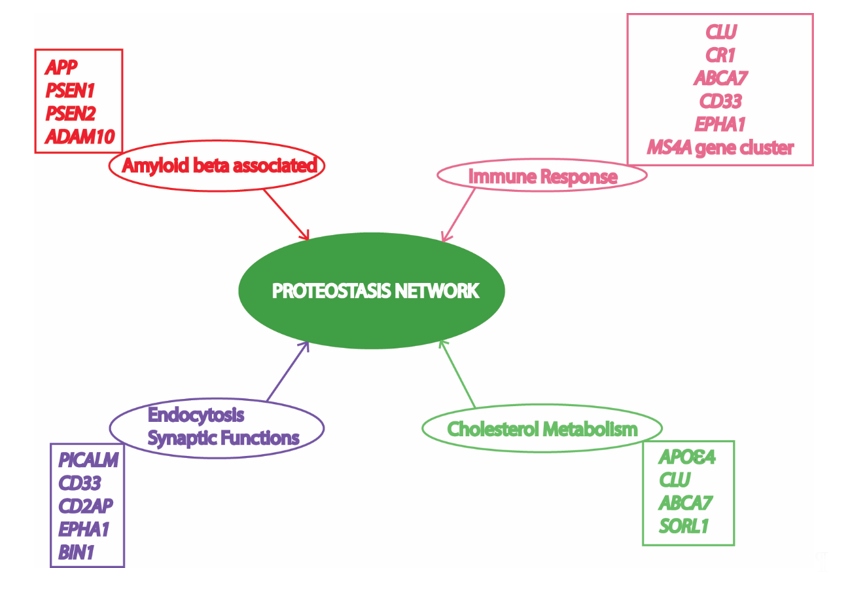
Alzheimer’s disease (AD) is an age-related progressive neurodegenerative disorder, which accounts for 60 to 80% deaths resulting from dementia in elderly people [1]. AD has been found to be a multifactorial disease with several causes being hypothesized and investigated [2]. However, amyloid beta (Aβ) mediated toxicity and its clearance from the neuronal environment has been the most popular area of AD research for the last three decades, as amyloid plaques are the most significant pathological hallmarks of the disease [3]. Memory deficits and cognitive impairment due to loss of neurons resulting in cortical atrophy accompanied by presence of amyloid plaques and neurofibrillary tangles are present in people with AD [4][5][4,5]. Depending upon the time of occurrence of first symptoms, AD victims have been classified into early onset AD (EOAD) and late onset AD (LOAD). Most AD patients (90–98%) have late onset disease with symptoms appearing after the age of 65 years [5]. AD pathology has been divided into familial autosomal dominant inheritance forms (<1% of total AD cases) and a sporadic multifactorial category (majority of cases) [6]. The mutations in amyloid precursor protein (APP) and two presenilin genes (PSEN1 and PSEN2) have been found to be associated with the familial form [7]. On the contrary, the multifactorial sporadic disease is more genetically complex, and cannot be attributed by any single risk factor or cause. More than 20 risk loci have been identified from previous studies involving genome wide association studies (GWAS) and linkage analysis (Figure 1). Most of these important AD-associated risk loci are associated with Aβ mediated toxic pathways, while other risk loci are associated with immune function, endocytosis, synapsis, and lipid metabolism [8]. Interestingly, all these cellular processes associated with AD pathology are somehow dependent on cellular protein homeostasis [9][10][11][12][13][9–13].
Figure 1. Risk factors/loci and associated cellular processes identified by genome wide association studies (GWAS) involved in Alzheimer’s Disease (AD) converge to the proteostasis network. APP, amyloid precursor protein; PSEN1, presenilin 1; PSEN2, presenilin 2; ADAM10, ADAM metallopeptidase domain 10; CLU, clusterin; CR1, complement C3b/C4b receptor 1; ABCA7, ATP binding cassette subfamily A member 7; CD33, CD33 molecule; MS4A, membrane spanning 4-domains A; PICALM, phosphatidylinositol binding clathrin assembly protein; CD2AP, CD2 associated protein; EPHA1, EPH receptor A1; BIN1, bridging integrator 1; APOε4, apolipoprotein E4; SORL1, sortilin related receptor 1. Various risk loci have been identified by GWAS to be associated with AD. Some of these risk loci are clustered to important cellular processes in this figure to depict their relationship with the proteostasis network, highlighting its significance in AD pathology. Previous studies suggest an important role of the proteostasis network in cellular processes associated with AD, including amyloid formation pathway, immune response mechanisms, vesicle trafficking, endocytosis, and lipid metabolism.
The intracellular neuronal environment of AD patients is characterized by loss of proteostasis, mitochondrial dysfunction, impaired mitophagy, oxidative damage, genetic instability, impaired protein clearance, amyloidosis, loss of synapsis, disruption of lipid homeostasis, biometal distribution alteration, and energy failure [2]. Most of these defects are found associated with various events in the AD brain; however, the majority are thought to be the consequence of Aβ overproduction and aggregation [14]. These intracellular cues are also conserved in yeast cells with more than 60% of its genes having human homologs or at least one conserved domain [15]. The major yeast models used for AD studies involve Saccharomyces cerevisiae [16]. These models for AD studies have been very important in understanding the role of conserved fundamental eukaryotic processes in AD pathology to develop methods to find potential therapeutics [17][18][19][20][21][22][23][17–23]. The conservation of signal transduction, energy metabolism, proteostasis network, lipid metabolism, vesicle trafficking, oxidative phosphorylation, stress response, longevity, and cell death in yeast models makes it highly appropriate for studies on AD pathology and treatment [15][24][15,24]. Various yeast models have been used in the past to understand AD progression and to find interventions to prevent and cure the disease [25].
2. Protein Homeostasis Network in Yeast
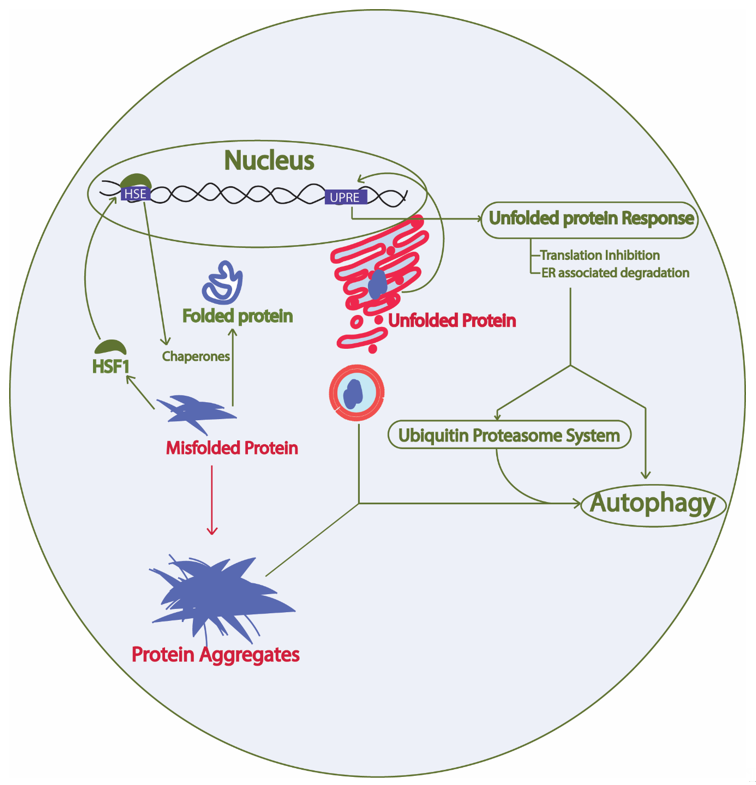
Constant turnover of cellular components inside a cell is of the utmost importance for adaptation during changing cellular environment, which interplays with multiple cellular processes required for cell survival [26]. Numerous proteins are coordinately functioning in a cell to fulfill the survival requirements of the cell. Protein synthesis, proper folding, protein quality control, transport of the protein to the target location, and proper function of the protein in its target depends on the cellular environment. Defects in protein processing or folding can cause abnormal aggregation of proteins possibly leading to disease if not repaired. These aberrations are detected by the cell and repaired by the protein quality control mechanisms governed by the proteostasis network. In fact, at least two types of chaperones in a eukaryotic system are controlling the protein folding: chaperones linked to protein synthesis (CLIPS) and chaperones expressed during stress [27]. In eukaryotes, chaperone mediated protein disaggregation machinery allows disaggregation of the protein aggregates and helps to correct the misfolding or partial unfolding of the protein [28]. However, failure to fold the proteins in correct conformation and inability of the chaperones to refold the aggregated proteins leads to activation of protein clearance mechanisms. Generally, protein clearance in the eukaryotic cell is controlled by three major molecular pathways: unfolded protein response, ubiquitin proteasome system, and autophagy (Figure 2) [29]. Various intrinsic and extrinsic factors play a crucial role in the regulation of these pathways inside a cell. Ageing has been found to be one important reason for the loss of proteostasis and subsequent disease development in neuronal cells due to accumulation of unwanted misfolded proteins [30]. During AD pathology, deposits of amyloid plaques and neurofibrillary tau tangles are the evidences that support the loss of protein homeostasis as leading cause of the disease [31]. Restoring the protein homeostasis has been hypothesized to cure or prevent AD and similar diseases [32]. This review elaborates on molecular mechanisms of unfolded protein response, autophagy, and ubiquitin proteasome system in yeast and their relevance to study human counterparts in relation to AD progression. Furthermore, we suggest possible applications of the knowledge of proteostasis network originating from yeast to find ways to explore AD pathology and develop drugs.
Figure 2. Proteostasis network involves protein synthesis, protein folding, heat shock response, unfolded protein response, ubiquitin proteasome system, and autophagy. Green arrow, protective; Red arrow, toxic; HSF1, heat shock factor 1; HSE, heat shock elements; UPRE, unfolded protein response elements. The proteostasis network involves protein synthesis machinery, protein folding, protein quality control, protein transport, and the overall turnover of proteins inside a cell. The presence of misfolded proteins in the cytosol trigger HSF1 activation, which activates promoters with HSE and results in expression of heat shock proteins that are either involved in protein disaggregation and refolding or clearance of the unwanted proteins through protein quality control mechanisms. Similarly, the presence of aberrations in the ER lumen causing unfolded protein formation leads to activation of the unfolded protein response and downstream activation of the protein quality control system.
2.1. Unfolded Protein Response Conserved in Yeast
Production of a protein is not sufficient for the proper biological function of protein; the expressed protein needs to be folded in its native form to be functional [33]. Most soluble proteins are folded directly in cytoplasm with the help of molecular chaperones [34], while the secreted proteins including hydrophobic membrane proteins pass through the endoplasmic reticulum (ER) and Golgi complexes and require correct folding [35]. Correct folding and maturity of the protein depends highly on the nutrient conditions, ER calcium homeostasis, availability of molecular chaperones, redox status, and cell health [36]. However, failure to maintain cellular homeostasis results in unfolded and misfolded proteins that are prone to aggregation. The formation of unfolded and misfolded proteins in the ER lumen triggers proteotoxic stress and activates the stress response pathway—the unfolded protein response (UPR) [37]. UPR is a mechanism by which the cells reduce the proteotoxic stress generated in the ER during protein folding and processing [36]. At least two types of stress response are activated during such conditions in mammalian cells. At first, the UPR attenuates protein translational activity by phosphorylating the translational initiation factor 2α (eIF2α) and activating the mRNA decay mechanism [38]. On the other hand, it increases protein folding, and activates autophagy and the ubiquitin proteasome system for degradation of undesired protein products [39]. On the other hand, UPR also activates the oxidative stress response to aid cell survival under ER stress [40].
2.1.1. Yeast Unfolded Protein Response
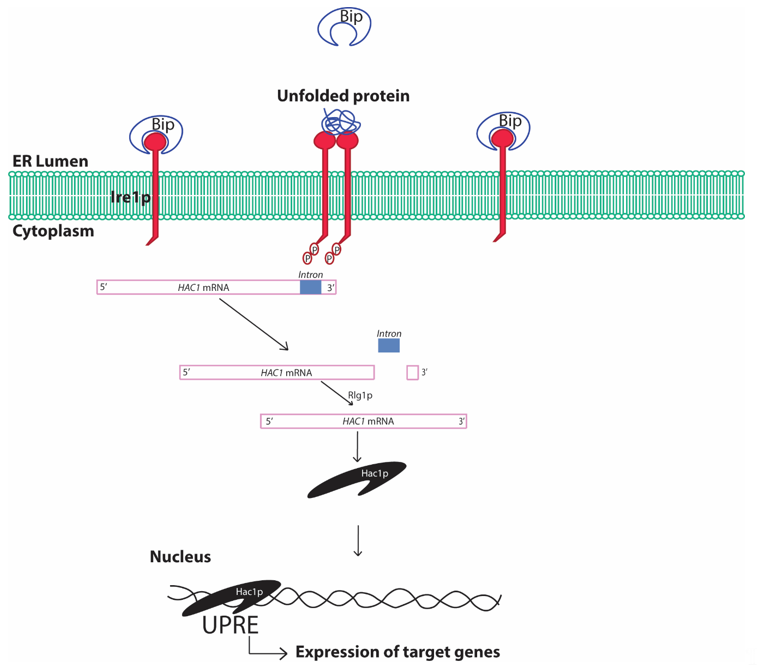
The UPR is conserved from the simplest eukaryotic unicellular yeast to complex human beings [36]. In yeast, the UPR involves sensing of unfolded proteins with the help of the stress sensor inositol requiring element 1 protein (Ire1p) in the ER lumen (Figure 3). The Ire1p is an ER localized type I transmembrane protein with its luminal domain sensing the protein folding environment in the ER and its cytosolic part comprising protein kinases and RNases activity. The luminal portion of the Ire1p is constantly bound with the chaperone binding immunoglobulin protein (Bip/GRP78) in normal conditions hindering the activation of Ire1p [41]. Any aberrations in the protein folding environment is indicated by the presence of unfolded and misfolded proteins in the ER or disassociation of Bip from Ire1p luminal part. The binding of Bip with Ire1p plays a crucial role in regulating UPR in yeast [42]. The oligomerization of Ire1p followed by its autophosphorylation is an essential activation signaling process that activates the cytosolic Ire1p RNase activity. The activation of Ire1p RNase activity leads to HAC1 (homologous to ATF/CREB1) mRNA splicing [43]. In normal conditions, HAC1 mRNA contains an inhibitory intron at the 3′ end, which renders HAC1 mRNA unfavorable for translation and remains inhibited by yeast protein two 1 (Ypt1p) [44]. Upon ER stress, the RNase activity of cytosolic part of Ire1p cleaves the specific inhibitory intron present in the 3′ end of the HAC1 mRNA. Moreover, re-joining of the cleaved active HAC1 mRNA ends is accomplished by tRNA ligase (Rlg1p) protein [45]. This processing of the HAC1 mRNA leads to its higher translation to Hac1p protein. Hac1p, protein encoded by the translationally active HAC1 mRNA, is a basic leucine zipper (bZIP) transcription factor that recognizes specific unfolded protein response elements (UPRE) sequences (5′-CACCTTG), present in the promoters of target genes including the Hac1 gene. Although Hac1p is sufficient for activation of UPR in yeast, complex interaction with other transcription factors involving interplay with Hac1p may result in activation of the UPRE promoters [46]. The upregulation of the promoters with UPRE region and subsequent UPR are protective to cells leading to the degradation of the unfolded protein by activating ER associated degradation (ERAD) [47]. The expression of stress resistance genes, autophagy genes and restoration of the energy metabolism further enhances cell survival. In addition, ER stress also reduces protein synthesis by inhibiting the eukaryotic translation initiation factor, which has been reported to be involved in assembly of mRNA-protein (RNP) granules [48]. At least two types of RNP granules, namely processing bodies (P-bodies) and stress granules, are found to be produced during various stresses including ER stress in yeast [49]. Formation of these granules, specifically the stress granules, are typically cytoprotective in nature and is an integral part of protein quality control.
Figure 3. Schematic diagram showing unfolded protein response in yeast cells. Bip, binding immunoglobulin protein; ER, endoplasmic reticulum; Ire1, inositol requiring element 1; HAC1, Homologous to ATF/CREB1; Rlg1p, tRNA ligase protein; UPRE, unfolded protein response elements. The presence of unfolded/misfolded proteins in the ER lumen is recognized by the yeast Ire1 sensor protein that activates the specific cytosolic endonuclease activity cleaving the inhibitory intron of HAC1 mRNA and rendering HAC1 mRNA translationally active. Hac1p protein translates and translocates to the nucleus, where it recognizes the characteristic cis-acting elements in the promoter regions of certain genes referred to as UPRE and increases expression of genes under control of such promoters. The process is referred to as the UPR.
2.1.2. Human Unfolded Protein Response
The human homologs of Bip, Ire1p, and Hac1p share high similarity in their molecular mechanism and the activation of important cytoprotective processes in mammalian cells, providing a rationale to explore yeast as model organism to study UPR and ER stress (Table 1). However, the mammalian UPR is more complex than the yeast UPR. The mammalian UPR is accomplished by three mechanisms dependent on three sensors, namely, inositol requiring element 1 (IRE1), PKR-like ER kinase (PERK), and activating transcription factor (ATF6), in the ER lumen [50]. The conserved IRE1 signaling in mammals causes the splicing of X-box binding protein 1 (XBP1, the Hac1p functional homolog in mammals) transcription factor mRNA and downstream signaling [51]. Additionally, the PERK protein is similar to Ire1p of yeast in the luminal part, while it is different in the cytosolic part. The activation of PERK leads to activation of the protein kinase domain that phosphorylates the eukaryotic translation initiation factor 2 alpha (eIF2α), which reduces the protein translational activity [52]. In humans as well, the inhibition of protein translation is associated with the formation of RNP granules, which is generally cleared by either protein disaggregation machinery or by autophagy [49]. In contrast, ATF6 sensors are absent in yeast and function very differently from the conventional response. Upon ER stress, ATF6p is transported to Golgi complexes, where it is cleaved by two proteases, specificity protein 1 (SP1) and specificity protein 2 (SP2), to release a fragment ATF6f. The ATF6f fragment translocates to the nucleus and acts as a bZIP transcription factor, which binds to the DNA in the promoter region of the target genes involved in ER associated degradation (ERAD) [52]. Meanwhile, the failure to protect cells from chronic ER stress may result in the activation of activating transcription factor 4 (ATF4)/C/EBP homologous protein (CHOP) signaling. The ER stress-induced CHOP signaling activates proapoptotic genes and reduces antiapoptotic gene expression [53]. Simultaneously, CHOP signaling also upregulates expression of growth arrest and DNA damage 34 (GADD34p) protein phosphatase, which dephosphorylates eIF2α enhancing protein synthesis, thereby inducing formation of reactive oxygen species as a result of compromised ER folding environment and chronic ER stress [54][55][54,55]. The overall effect of chronic ER stress is characterized by the activation of programmed cell death via apoptosis [56].
Table 1.
Conserved proteins of yeast unfolded protein response and their human homologs
.
|
Yeast Protein |
Human Homolog |
Function |
References |
|
Kar2p/Bip/Grp78p |
HSPA5/BiP/GRP78 |
Chaperone regulating activation of Ire1p |
|
|
Ire1p |
IRE1α IRE1β |
Stress sensing and endonuclease activity |
[58] |
|
PERK |
Stress sensing and protein kinase |
[59] |
|
|
Hac1p |
XBP1p |
UPRE binding and expression of target genes |
[59] |
|
Sui2p |
eIF2α |
Eukaryotic translation initiation factor involved in protein translation regulation |
[60] |
|
Ypt1p |
RAB1Ap |
Regulates UPR by HAC1 mRNA decay |
[44] |
|
Yeast Protein |
Human Homolog |
Function |
References |
|
Kar2p/Bip/Grp78p |
HSPA5/BiP/GRP78 |
Chaperone regulating activation of Ire1p |
[42,57] |
|
Ire1p |
IRE1α IRE1β |
Stress sensing and endonuclease activity |
[58] |
|
PERK |
Stress sensing and protein kinase |
[59] |
|
|
Hac1p |
XBP1p |
UPRE binding and expression of target genes |
[59] |
|
Sui2p |
eIF2α |
Eukaryotic translation initiation factor involved in protein translation regulation |
[60] |
|
Ypt1p |
RAB1Ap |
Regulates UPR by HAC1 mRNA decay |
[44] |
References
- Alzheimer's 2020 Alzheimer's disease facts and figures. Alzheimer's Dement. 2020, 16, 391–460.
- Dhakal, ; Kushairi, N.; Phan, C.W.; Adhikari, B.; Sabaratnam, V.; Macreadie, I. Dietary polyphenols: A multifactorial strategy to target Alzheimer’s disease. Int. J. Mol. Sci. 2019, 20, 5090.
- Panza, ; Lozupone, M.; Logroscino, G.; Imbimbo, B.P. A critical appraisal of amyloid-β-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 2019, 15, 73–88.
- Braak, ; Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991, 82, 239–259.
- Cuyvers, ; Sleegers, K. Genetic variations underlying Alzheimer's disease: Evidence from genome-wide association studies and beyond. Lancet. Neurol. 2016, 15, 857–868.
- Brouwers, ; Sleegers, K.; Van Broeckhoven, C. Molecular genetics of Alzheimer's disease: An update. Ann. Med. 2008, 40, 562–583.
- Bertram, ; Tanzi, R.E. Alzheimer disease risk genes: 29 and counting. Nat. Rev. Neurol. 2019, 15, 191–192.
- Bettens, ; Sleegers, K.; Van Broeckhoven, C. Genetic insights in Alzheimer's disease. Lancet Neurol. 2013, 12, 92–104.
- Xie, ; Li, J.; Kang, R.; Tang, D. Interplay between lipid metabolism and autophagy. Front Cell Dev Biol 2020, 8, 431–431.
- Birgisdottir, Å.B.; Johansen, Autophagy and endocytosis – interconnections and interdependencies. J. Cell Sci. 2020, 133, jcs228114.
- Nikoletopoulou, ; Tavernarakis, N. Regulation and roles of autophagy at synapses. Trends Cell Biol. 2018, 28, 646–661.
- Reddy, P.H.; Oliver, D.M. Amyloid beta and phosphorylated tau-induced defective autophagy and mitophagy in Alzheimer’s Cells 2019, 8, 488.
- Loureiro, ; Ploegh, H.L. Antigen presentation and the ubiquitin—Proteasome system in host–pathogen interactions. In Advances in Immunology; Academic Press: Cambridge, MA, USA, 2006; Volume 92, pp. 225–305.
- LaFerla, F.M.; Green, K.N.; Oddo, Intracellular amyloid-β in Alzheimer's disease. Nat. Rev. Neurosci. 2007, 8, 499–509.
- Khurana, ; Lindquist, S. Modelling neurodegeneration in Saccharomyces cerevisiae: Why cook with baker's yeast? Nat. Rev. Neurosci. 2010, 11, 436–449.
- Seynnaeve, ; Vecchio, M.D.; Fruhmann, G.; Verelst, J.; Cools, M.; Beckers, J.; Mulvihill, D.P.; Winderickx, J.; Franssens, V. Recent insights on Alzheimer's disease originating from yeast models. Int. J. Mol. Sci. 2018, 19, 1947.
- Caine, ; Sankovich, S.; Antony, H.; Waddington, L.; Macreadie, P.; Varghese, J.; Macreadie, I. Alzheimer's Abeta fused to green fluorescent protein induces growth stress and a heat shock response. FEMS Yeast Res. 2007, 7, 1230–1236.
- Porzoor, ; Alford, B.; Hügel, H.M.; Grando, D.; Caine, J.; Macreadie, I. Anti-amyloidogenic properties of some phenolic compounds. Biomolecules 2015, 5, 505–527.
- Chen, ; Petranovic, D. Amyloid-β peptide-induced cytotoxicity and mitochondrial dysfunction in yeast. FEMS Yeast Res. 2015, 15, fov061.
- Chen, ; Bisschops, M.M.M.; Agarwal, N.R.; Ji, B.; Shanmugavel, K.P.; Petranovic, D. Interplay of energetics and ER stress exacerbates Alzheimer's amyloid-β (Aβ) toxicity in yeast. Front. Mol. Neurosci. 2017, 10, 232.
- Chen, ; Ji, B.; Hao, X.; Li, X.; Eisele, F.; Nyström, T.; Petranovic, D. FMN reduces Amyloid-β toxicity in yeast by regulating redox status and cellular metabolism. Nat. Commun. 2020, 11, 867.
- Bharadwaj, P.R.; Martins, R.N. Autophagy modulates Aβ accumulation and formation of aggregates in Mol. Cell. Neurosci. 2020, 104, 103466.
- Macreadie, I.G.; Arvanitis, ; Bharadwaj, P. Finding chemopreventatives to reduce amyloid beta in yeast. Neural Regen Res. 2016, 11, 244–245.
- Macreadie, ; Dhakal, S. Insights from yeast on oxidative stress in Alzheimer’s disease, focusing on Ahp1p/Prx5. OBM Geriartrics 2019, 3, 10.
- Mcdonald, J.B.; Dhakal, ; Macreadie, I.G. Yeast contributions to Alzheimer’s Disease. J. Human. Clin. Gen. 2020, 2, 1–19.
- Webster, B.M.; Gildea, H.K.; Dillin, Protein homeostasis from the outside in. Nat. Cell Biol. 2020.
- Liberek, ; Lewandowska, A.; Zietkiewicz, S. Chaperones in control of protein disaggregation. Embo J. 2008, 27, 328–335.
- Verghese, ; Abrams, J.; Wang, Y.; Morano, K.A. Biology of the heat shock response and protein chaperones: Budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol. Mol. Biol. Rev. 2012, 76, 115–158.
- Hartl, F.U.; Bracher, ; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332.
- Hipp, M.S.; Kasturi, ; Hartl, F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019, 20, 421–435.
- Morawe, ; Hiebel, C.; Kern, A.; Behl, C. Protein homeostasis, aging and Alzheimer's disease. Mol. Neurobiol. 2012, 46, 41–54.
- Cheng, ; North, B.J.; Zhang, T.; Dai, X.; Tao, K.; Guo, J.; Wei, W. The emerging roles of protein homeostasis-governing pathways in Alzheimer's disease. Aging Cell 2018, 17, e12801–e12801.
- Englander, S.W.; Mayne, The nature of protein folding pathways. Proc. Natl. Acad. Sci. USA 2014, 111, 15873.
- Vabulas, R.M.; Raychaudhuri, ; Hayer-Hartl, M.; Hartl, F.U. Protein folding in the cytoplasm and the heat shock response. Cold Spring. Harb. Perspect Biol. 2010, 2, a004390–a004390.
- Skach, W.R. Cellular mechanisms of membrane protein Nat. Struct Mol. Biol. 2009, 16, 606–612.
- Hetz, The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012, 13, 89–102.
- Mori, Signalling Pathways in the unfolded protein response: Development from yeast to mammals. J. Biochem. 2009, 146, 743–750.
- Chakrabarti, ; Chen, A.W.; Varner, J.D. A review of the mammalian unfolded protein response. Biotechnol. Bioeng. 2011, 108, 2777–2793.
- Schröder, ; Kaufman, R.J. ER stress and the unfolded protein response. Mutat. Res. /Fundam. Mol. Mech. Mutagenesis 2005, 569, 29–63.
- Guerra-Moreno, ; Ang, J.; Welsch, H.; Jochem, M.; Hanna, J. Regulation of the unfolded protein response in yeast by oxidative stress. FEBS Lett. 2019, 593, 1080–1088.
- Wu, ; Ng, B.S.; Thibault, G. Endoplasmic reticulum stress response in yeast and humans. Biosci. Rep. 2014, 34, e00118.
- Okamura, ; Kimata, Y.; Higashio, H.; Tsuru, A.; Kohno, K. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem. Biophys. Res. Commun. 2000, 279, 445–450.
- Sidrauski, ; Walter, P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 1997, 90, 1031–1039.
- Tsvetanova, N.G.; Riordan, D.P.; Brown, P.O. The yeast rab GTPase Ypt1 modulates unfolded protein response dynamics by regulating the stability of HAC1 PLoS Genet. 2012, 8, e1002862.
- Mori, ; Ogasawara, C.; Inada, T.; Englert, M.; Beier, H.; Takezawa, M.; Endo, T.; Yoshihisa, T. Dual functions of yeast tRNA ligase in the unfolded protein response: Unconventional cytoplasmic splicing of HAC1 pre-mRNA is not sufficient to release translational attenuation. Mol. Biol. Cell 2010, 21, 3722–3734.
- Ogawa, ; Mori, K. Autoregulation of the HAC1 gene is required for sustained activation of the yeast unfolded protein response. Genes Cells 2004, 9, 95–104.
- Hwang, ; Qi, L. Quality control in the endoplasmic reticulum: Crosstalk between ERAD and UPR pathways. Trends Biochem. Sci. 2018, 43, 593–605.
- Protter, D.S.W.; Parker, Principles and properties of stress granules. Trends Cell Biol. 2016, 26, 668–679.
- Buchan, J.R.; Parker, Eukaryotic stress granules: The ins and outs of translation. Mol. Cell 2009, 36, 932–941.
- Hetz, ; Zhang, K.; Kaufman, R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020, 21, 421–438.
- Plumb, ; Zhang, Z.-R.; Appathurai, S.; Mariappan, M. A functional link between the co-translational protein translocation pathway and the UPR. eLife 2015, 4, e07426.
- Adachi, ; Yamamoto, K.; Okada, T.; Yoshida, H.; Harada, A.; Mori, K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct. Funct. 2008, 33, 75-89.
- Galehdar, ; Swan, P.; Fuerth, B.; Callaghan, S.M.; Park, D.S.; Cregan, S.P. Neuronal apoptosis induced by endoplasmic reticulum stress is regulated by ATF4–CHOP-mediated induction of the Bcl-2 homology 3-only member PUMA. J. Neurosci. 2010, 30, 16938.
- Novoa, ; Zeng, H.; Harding, H.P.; Ron, D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J. Cell Biol. 2001, 153, 1011–1022.
- Landau, ; Kodali, V.K.; Malhotra, J.D.; Kaufman, R.J. Chapter Fourteen - Detection of oxidative damage in response to protein misfolding in the endoplasmic reticulum. In Methods in Enzymology; Cadenas, E., Packer, L., Eds.; Academic Press: Cambridge, MA, USA, 2013; Volume 526, pp. 231–250.
- Tabas, ; Ron, D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 2011, 13, 184–190.
- Rose, M.D.; Misra, L.M.; Vogel, J.P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 Cell 1989, 57, 1211–1221.
- Zhang, ; Zhang, C.; Wang, A. Divergence and conservation of the major UPR branch IRE1-bZIP signaling pathway across eukaryotes. Sci. Rep. 2016, 6, 27362.
- Kohno, Stress-sensing mechanisms in the unfolded protein response: Similarities and differences between yeast and mammals. J. Biochem. 2010, 147, 27–33.
- Laurino, J.P.; Thompson, G.M.; Pacheco, ; Castilho, B.A. The beta subunit of eukaryotic translation initiation factor 2 binds mRNA through the lysine repeats and a region comprising the C2-C2 motif. Mol. Cell. Biol. 1999, 19, 173–181.