Brassica vegetables are very important to human beings. Self-incompatibility (SI) is a common phenomenon in Brassica. Breeding by SI lines is an important way to utilize heterosis of Brassica vegetables. It is believed that the SI inheritance in Brassica species is controlled by three linkage genes on the S-locus, including SRK (S-locus receptor kinase), SCR (S-locus cystine-rich protein)/SP11 (S-locus protein 11), and SLG (S-locus glycoprotein). SRK is the female determinant and SCR/SP11 is the pollen S gene. The expression of SLG is necessary for SRK, and it enhances the SRK-mediated SI reaction. In addition to these three S-locus genes, some other functional molecules also have significant regulatory effects on SI, such as ARC1 (arm repeat containing 1), MLPK (M-locus protein kinase), Exo70A1 (exocyst compounds), THLl/THL2 (thioredoxin H-like), MOD (aquaporin), SLR (S-locus-related glycoprotein), BPCI (pollen calcium-binding protein I), etc. SI is also associated with the dominant/recessive relationship between S alleles.
1. Introduction
Brassica vegetables, such as cabbage, Chinese cabbage, cauliflower, broccoli, etc., are important foods for people all over the world. Self-incompatibility (SI) is one of the main ways to utilize heterosis of Brassica vegetables. Using SI lines for breeding not only avoids an emasculation operation but also obtains heterosis. SI is widely used in the breeding of Brassica vegetables and has greatly improved the yield and quality of these vegetables. In China, ”Jingfeng No. 1” was the first cabbage variety to be developed using SI lines and was developed in 1973. Later, SI lines were used to breed Qingfeng, Muchun, Zhengchun, Wanfeng, 8398, and other varieties. Understanding the molecular mechanisms of the SI recognition reaction in Brassica vegetables is important for utilizing heterosis.
SI refers to a phenomenon that plants have complete flowers and can form normal female and male gametes, but cannot bear seeds through self-pollination or the rate of fruiting is very low. SI is a widespread mechanism that helps prevent detrimental inbreeding and maintains genetic variation within higher plants. According to the morphology of perfect flowers, SI can be classified as heteromorphic SI and homomorphic SI. Homomorphic SI occurs in species with uniform flower morphology and is further divided into gametophytic SI (GSI) and sporophytic SI (SSI)
[1]. GSI is controlled by the genotype of the gametophyte and SSI controlled by the genotype of pollen parents. SI in Brassica vegetables belongs to SSI and is sporophytically controlled by the S-locus. Three tightly linked genes are distributed on the S-locus, including SRK (S-locus receptor kinase), SLG (S-locus glycoprotein), and SCR (S-locus cysteine-rich protein)/SP11 (S-locus protein 11). The first two genes are exclusively expressed in the pistil and the last one is expressed in pollen grains
[2]. SRK and SCR/SP11 function as receptor–ligand pairs that determine specificity in the stigma epidermis and pollen
[3]. In addition, many other elements also affect SI, such as ARC1 (Aarm repeat containing 1), MLPK (M-locus protein kinase), Exo70A1 (exocyst compounds), THLl/THL2 (thioredoxin H-like), MOD (aquaporin), SLR (S-locus-related glycoprotein), BPCI (pollen calcium-binding protein I), etc.
2. S-Locus Transducers of SSI
2.1. SRK
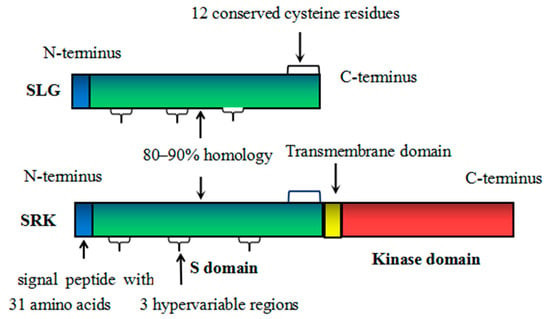
SRK was the receptor that allowed stigma epidermal cells to discriminate between genetically self- and non-self-pollen in SI responses of Brassicaceae
[2]. SRK encoded a trans-membrane protein which contained an S domain, a trans-membrane region, and a cytoplasmic serine/threonine (Ser/Thr) protein kinase domain (
Figure 1), and it was proposed to be the female determinant of SI reaction
[4]. The SRK gene of
Brassica oleracea contained seven exons and six introns, and introns varied in size from 76 bp to 896 bp. The derived protein products contained 857 amino acids. Exon 1 encoded the S-region and was composed of 438 amino acids
[5]. Its N-terminal contained a signal peptide consisting of 31 hydrophobic amino acids with 67% homologous to SLG. The remaining S-domain contained 12 almost completely conserved cysteine residues and three polytropic regions, with 89% homologous to SLG. The conservative cysteines played an important role in SRK function. The trans-membrane domain was encoded by exon 2 and contained 20 hydrophobic amino acids. The exon 3–7 encoded protein kinase domain, which was the functional site and was used to express and transmit signals from the extracellular domain through phosphorylation of Ser and Thr
[5]. The SRK extracellular ligand-binding domain contained several potential N-glycosylation sites. In Arabidopsis, although five of six potential N-glycosylation sites in SRKb were glycosylated in stigma, N-glycosylation was not important for SCRb-dependent activation of SRKb. N-glycosylation functioned primarily to ensure the proper and efficient sub-cellular trafficking of SRK to the plasma membrane
[6].
Figure 1. Schematic structure of SLG and SRK of Brassica vegetables. The signal peptide is colored blue. The S domain is colored green. The transmembrane domain is colored yellow. The kinase domain is colored red. SRK consists of an S domain, a transmembrane domain, and a kinase domain. Both SRK and SLG have a signal peptide with 31 amino acids, 3 hypervariable regions and 12 cysteine residues. SRK is similar to SLG in the S domain.
SRK was specifically expressed in the stigma, whose expression level was closely related to SI
[7]. Takasaki et al. introduced the SRK
28 gene of S haplotype (S
28) into S
60 of
Brassica rapa, and all the 17 transgenic plants obtained were SI. Among them, 3 plants rejected pollen of S
28 and S
60, while the other 14 only rejected S
60 pollen, which indicated that the stigma SI phenotype of the first 3 plants changed and differed from the parent S
60. Using pollen of S
24, S
43, S
45, S
52, and S
29 for pollination, they obtained a SI phenotype specific to SRK
28, with the SRK
28 transcript being detected in stigma but not in anther
[8]. Silva et al. introduced SRK
910 into SC (self-compatibility) of
Brassica napus and found that SI occurred
[9]. These evidences proved that SRK was not only a determinant of pistil, but also played an important role in pollen recognition. When homologous SCR fell and bound to SRK, its intracellular kinase area activated, Ser and Thr residues phosphorylated, resulting in a cascade reaction that transmitted and amplified signals in mastoid cells and ultimately inhibited pollen germination and pollen tube growth.
2.2. SCR/SP11
SCR/SP11 encoded a small cysteine-rich protein, secreted from the anther tapetum, localized to the pollen surface and functioned as the male determinant of SI reaction
[10,11][10][11]. In SC mutants of cabbage, SCR will not express, and as a result SI is lost. Analysis showed that SCR and SP11 was the same gene with a single copy, encoding an 8.4–8.6 kD small molecule protein
[12]. The protein of SCR was alkaline and hydrophilic, belonged to the pollen coat protein family, and accumulated and was specifically expressed in anthers. The SCR protein contained eight cysteine residues and one glycine residue, which were conserved in most Brassica SCR alleles
[13,14][13][14]. Analysis of the amino acid sequence of Brassica SCR of S
6, S
8, and S
13 of
B. oleracea showed that the similarity was only 30–42%. The high variation indicated that it had specific SI alleles. The pollen of transgenic plants had the S
6 pollen phenotype (constructed expression vectors with the SCR
8 promoter and SCR
6 gene, and transferred into S
2S
2 homozygous of cabbage), which verified that SCR was a pollen S gene
[12]. Analysis of SP11 transgenic plants of
B. rape showed that SI occurred when the pollen of the transgenic plants was pollinated to the stigma of the same haplotype. However, when it was pollinated to different haplotypes, SC happened
[14]. Takayama et al. expressed SP11 protein of
Brassica campestris in vitro, and then treated the same haplotype material stigma mastoid cells with this protein. The results showed that the hydration of pollen was inhibited and incompatibility happened, which also proved that SCR/SP11 was the S gene of pollen
[10]. In Brassica, the expression of SP11 can be suppressed by two small RNAs, SP11 methylation inducer (SMI1) and SMI2
[15]. SCR had high homology among some species. Haseyama et al. reported that the SCR of S
31 in cabbage, S
69, and S
52 in rape were highly homologous to those of S
22, S
25, and S
30 in radish with the amino acid identities being 84.6%, 87.7%, and 82.0%, respectively. However, the homology was low between intergeneric pairs
[16].
2.3. SLG
The first S-locus product to be identified was SLG, which encoded a secreted glycoprotein and was first isolated from cabbage. In some Brassica genotypes, the highest content of SLG was as much as 5% of the total stigma soluble protein
[17]. The accumulation of SLG on the stigma was significantly correlated with SI
[18]. The SLG was shown to increase during stigma development and to reach maximal levels simultaneously with the onset of the SI response
[19]. The molecular weight of SLG was 57–65 kD, distributed in the cell wall and intercellular region of the stigma papilla cells
[20]. The SLG proteins of different genotypes had different molecular weights. In
B. oleracea, it consisted of 436 amino acids. The functional area comprised 405 amino acid residues. The N-terminal (1–181 amino acids) was hydrophobic and highly conserved (approximately 80%) with a signal peptide of 31 amino acids, which was responsible for guiding SLG into the cellular secretion system. The residues of 182–268 amino acids showed the greatest variation, with only 52% conservative. The C-terminal (269–436 amino acids) contained 12 completely conserved cysteine residues and 12 N-glycosylation sites, with the C-terminal approximately 78% conserved
[20] (
Figure 1). The Asn and n-GLCNAC on the glycosylation sites of
Brassica oleracea were connected by N-glucoside bonds
[21]. The arrangement of N-glycation sites of different SLG proteins was different. The relationship between the change of this structure and the specific pairing of the S allele product was still unclear
[22]. Amino acid sequences of SLG was highly similar to SRK, with both having 12 conserved cysteine residues and three hypervariable regions
[1]. The homology between SLG and SRK was approximately 80–90% in
B. rapa,
B. oleracea, and
B. napus (
Figure 1)
[4]. It was once thought that SLG was only responsible for pollen adhesion, but now SLG diversity is interpreted as evidence of a role in SI
[23]. Furthermore, the degree of S pollen rejection by SRK was correlating with the degree of amino-acid identity in S domain between SRK and SLG; the higher the identity, the stronger the rejection
[8].
2.4. Genomic Organization of S-Locus Genes
The genomic organization and transcription direction of three S-locus genes were different among species. For example, in
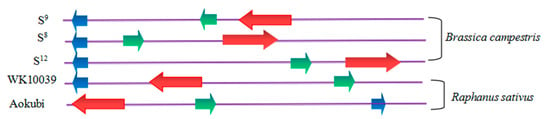
B. campestris S
8, S
9, and S
12, SCR/SP11 gene was located between SLG and SRK
[10]. In radish WK10039, the SRK gene was located between SCR/SP11 and SLG. However, in Aokubi, the SCR/SP11 gene was located between SLG and SRK
[24]. The direction of transcription of these three S-locus genes were divergent as shown in
Figure 2. The length of the S-locus in Brassica was usually 200–400 kb
[10,12][10][12]. However, the physical distance between SLG and SRK varied greatly, from as little as a few kb in
Brassica campestris (syn
B. rapa) to as much as several hundred kb in
B. oleracea. In cabbage, it was more than 200 kb in S
2 and S
6, less than 50 kb in S
13, and about 13 kb in rape S
8 [25]. It was believed that SRK and SLG may co-evolve through gene conversion. The co-evolution of SLG and SRK suggested that SLG was related to S haplotypes
[26]. In the case of
Brassica napus, the S-locus region was found to originate from the introgressed
Brassica rapa genome and not from
Brassica oleracea [4].
Figure 2. Schematic organization of S-locus genes. The arrow shaped boxes indicate S genes and the direction of transcription. The SLG is colored blue. The SCR/SP11 is colored green. The SRK is colored red. SCR/SP11 gene is located between SLG and SRK in Brassica campestris and Raphanus sativus Akokubi, while SRK gene is located between SLG and SCR/SP11 in Raphanus sativus WK10039. The direction of transcription of these genes were divergent in B. campestris and Raphanus sativus.
2.5. The Relationship of S-Locus Genes in SI Reaction
During self-pollination, pistil factor SRK localized on the plasma membrane of the papilla cell, specifically recognized its cognate pollen factor SCR/SP11 which was released from the pollen surface, triggered self-phosphorylation of the kinase domain and transduction the SI signal to intracellular downstream effector. SLG can form a complex with SCR. Then, the signal peptide guided the SCR-SLG complex to pass through the intercellular space and reach the cell wall of the stigma epidermal cell. Here, the signal peptide dissociated, the complex conformation changed, and the SCR ligand bound specifically to the extracellular domain of the homologous SRK in the stigma and activated intracellular kinase. The SRK conformation was affected by SLG which functioned in the stabilization and proper processing of SRK
[1]. The expression of SLG was necessary for the physiological activity of SRK. SLG and SRK must be expressed together, otherwise SRK forms a polymer with high molecular weight. SRK and SLG may interact through the same receptor or ligand
[27,28][27][28]. How did SRK discriminate self or non-self SCR/SP11? Murase et al. established a model of self/non-self-discrimination in Brassica. They thought that the binding free energies were most stable for cognate SRK-SP11 combinations, and the modes of SRK–SP11 interactions differed between intra- and inter-subgroup combinations
[15].