Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Venugopal Rao Soma and Version 2 by Camila Xu.
Surface-enhanced Raman spectroscopy/scattering (SERS) has evolved into a popular tool for applications in biology and medicine owing to its ease-of-use, non-destructive, and label-free approach. Advances in plasmonics and instrumentation have enabled the realization of SERS’s full potential for the trace detection of biomolecules, disease diagnostics, and monitoring.
- biosensing
- SERS
- plasmonics
- disease diagnosis
1. Introduction
Plasmonics is the study of electron oscillations in metal nanostructures and their interaction with electromagnetic radiation. Since its conception in the 1950s, researchers have been interested in studying the fundamentals of the effects of shape, surrounding medium, material, and their interaction with light of different wavelengths [1]. With this well-established knowledge, plasmonics is witnessing an enormous potential for applications in different fields, including forensics [2]; environmental safety [3]; biosensing [4][5][6][7][8][9][10][11][4,5,6,7,8,9,10,11], e.g., SARS-CoV-2 detection [12]; and homeland security [13]. The applications of plasmonics majorly rely on surface plasmon resonance (SPR) or localized surface plasmon resonance (LSPR) effects [14]. Some of the significant techniques that were developed using these include higher-order harmonic generation, microscopy, drug delivery, photovoltaics, surface-enhanced Raman spectroscopy (SERS) and fluorescence, and surface-enhanced infrared absorption spectroscopy (SEIAS) and waveguides. The use of plasmonics in these techniques has significantly improved their efficiency over existing conventional techniques, offering flexibility, signal enhancement, and ease of use [15]. Advents in plasmonics have led to the emergence of SERS with impressive signal enhancements over traditional Raman spectroscopy [16]. SERS-based sensing is being widely used for the trace detection of different molecules, such as explosives [17], pesticides [18][19][18,19], food adulterants [20][21][20,21], drugs [22], biomolecules [23][24][25][26][27][23,24,25,26,27], medicine [28][29][30][28,29,30], and microorganisms [31].
SERS typically utilizes localized surface plasmon resonances in metal nanostructures to enhance the weak Raman signal significantly. The phenomenon was first observed by Fleischmann in 1974 while studying pyridine adsorbed on a roughened silver electrode [32]. However, the enhancement was attributed to increased surface area for adsorption. It took further experiments in 1977 by two independent groups, Jeanmaire and van Duyne [33] and Albrecht and Creighton [34], to understand the origin of the enhancement. Now it is established that the enhancement predominantly comes from two mechanisms: electromagnetic enhancement (EE) and chemical enhancement (CE) [35]. The electromagnetic enhancement in SERS is a two-step process, and the total enhancement is multiplicative. When a molecule of interest is in the vicinity of a plasmonic nanostructure, it experiences an enhanced field called local field enhancement (LFE). The molecule then radiates with increased efficiency, referred to as radiation enhancement [36][37][36,37]. In addition, there is chemical enhancement which occurs because of charge-transfer mechanisms between nanoparticles and the analyte. Figure 1 summarizes the two enhancement mechanisms in SERS. The type of the plasmonic material, choice of wavelength, surface coverage of the molecules, and concentration of the analyte are the factors that influence SERS’s efficiency [38]. This technique is label-free, rapid, non-destructive, and water compatible and offers the fingerprint of the molecule, making it suitable for numerous applications. Nobel metals such as Au, Ag, and Cu and their alloys are the widely used materials for SERS for their tunability in the visible and IR region, inertness, sensitivity, and compatibility [39][40][39,40]. Despite the superior performance of Ag owing to its high-quality resonance in the visible region, Au is the preferred material, as it is known to be biocompatible and non-reactive in an oxygen atmosphere. The near-field enhancement in SERS is dependent on the shape and size of the nanostructures, in addition to the distance between the nanoparticles and distribution of probe molecules around the nanoparticles [41]. The different morphologies of nanoparticles, such as core–shell, rods, spherical, triangular, stars, and nanopyramids, are synthesized by widely reported chemical routes in bottom-up or top-down approaches [42]. Anisotropic nanostructures such as dendrites, rods, stars, and triangle are considered highly desirable for SERS since they enable lower detection limits owing to the lightening-rod effect [43][44][43,44]. The performance of SERS is also dependent on the choice of wavelength, and most biological tissues are transparent in the IR region, making it a preferred choice [45]. Recently, there is also growing interest in the UV and deep UV SERS for applications concerning biomolecules such as amino acids and DNA bases because they have electronic transitions in the UV region [39].
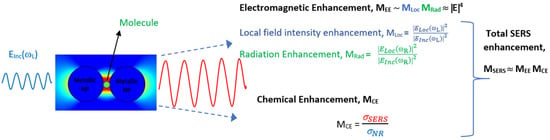
Figure 1.
Schematic of total enhancement in SERS via electromagnetic and chemical enhancement mechanisms.
With a growing population and, consequently, the diseases worldwide, there is a need to develop point-of-care (POC) devices that are easy to use, reliable, rapid, and low cost. Over the years, SERS has been proven to possess all of these advantages, including trace detection with sub-picomolar sensitivity. Particularly, there are many reasons for the surge of using SERS for biosensing. Firstly, given the low scattering cross-section of water, SERS is extremely compatible with liquid samples, paving the way for use in biology applications, including liquid biopsy [46][47][46,47]. SERS has been widely used for disease diagnosis using urine, blood, serum, plasma, saliva, breath, and tear samples, establishing its compatibility. Measurements in SERS can be performed using liquids, gases, solids, and powders, unlike traditional tests. Secondly, SERS gives specific molecular information, which is often a vibrational fingerprint of the molecule or cell under study. Biomarkers that are Raman active are extensively used for the identification of different diseases, using SERS [48]. Frequently, when the variations are unrecognizable to the human eye, machine learning techniques are used to extract the patterns and discriminate the samples [49]. This was successfully used to classify normal and cancer cells [50], identify microorganism species [51], and monitor disease progression [52]. Thirdly, SERS is a rapid technique, can accomplish trace detection, and has a test time of three to five minutes [53]. Combined with recent developments in flexible SERS sensors, it also offers easy sample-collection methods, such as swabbing from an uneven surface [54]. Lastly, advances in portable instrumentation and low-cost lasers leveraged the usage of SERS for real-world applications [55]. The easy availability of IR lasers that have a low damage threshold with biology samples, as well as quench fluorescence, has favored the development of SERS for biosensing. All of these advantages have made SERS a popular choice for biosensing recently.
2. SERS for Disease Diagnosis
With growing zoonotic diseases, cancers, diabetes, and other ailments, there is a pressing need to develop low-cost and POC identification techniques. Early and rapid diagnosis is the key to saving a life and prevent the rapid transmission of diseases. A trace detection technique such as SERS will aid in tracking the minute changes in cells or biomarkers, thus enabling early diagnosis. SERS is being extensively used for the same in both labeled and label-free approaches, often targeting specific biomarkers of the disease expression [30]. In the label-free approach, the sample is directly studied in contact with the plasmonic material, whereas in the labeled approach, a Raman reporter, such as fluorophores, antibodies, or ligands, is attached to the sample for detection and imaging [56][57][66,67]. Different biomarkers, such as proteins, antibodies, miRNAs, exosomes, and DNA, are used as indicators for the presence of the disease. In researchers observation, where full cells, tissues, or body fluids are studied, a machine learning algorithm is used hand in hand for accurate identification. SERS has been used for the detection of conditions such as Alzheimer’s [58][59][60][61][68,69,70,71], PCOS [62][72], diabetes [63][64][73,74], inflammation [64][74], Crohn’s disease [65][75], and single Hb molecule [66][76], to name a few.2.1. Cancer Diagnosis and Theranostics
Cancer is the new pandemic and a leading cause of deaths in the modern world [67][77]. There is an increase in the incidence of various types of cancers, including mouth, gastric, lungs, ovaries, skin, and blood cancer. Numerous factors, such as environment, diet, lifestyle, and smoking, can trigger cancer. The early diagnosis of cancer is extremely important, as it is lifesaving with existing treatment protocols. Conventional cancer diagnosis is often performed using imaging techniques such as X-ray, computerized tomography scan (CT), positron emission tomography (PET), ultrasound, and magnetic resonance imaging (MRI). These techniques are often destructive, posing the risk of radiation ionization, and are often not compatible with patients with pre-existing conditions and medical devices such as pacemakers [68][78]. These are also expensive, involve sophisticated instruments, are time-consuming, and are often performed with multiple tests to avoid ambiguity [69][79]. Recently, there has been an increase in using plasmonic biosensing for cancer diagnosis and therapy, with review articles summarizing the progress in the same [70][71][72][73][74][75][76][80,81,82,83,84,85,86]. They are established to be minimally invasive, rapid, low cost, and offer point-of-care testing [77][78][87,88]. Of all plasmonic-based detection techniques, SERS is being extensively used for cancer identification, monitoring, and other theranostics, including imaging and chemo/photothermal therapy [79][80][81][82][83][84][85][86][89,90,91,92,93,94,95,96]. SERS facilitates liquid biopsy [86][96] by using urine, saliva, and serum, thus making it low cost and enabling easier frequent sampling compared to the existing tissue-biopsy techniques, which are often destructive [87][97]. Different cancer biomarkers, such as miRNA [88][89][98,99], proteins, exosomes [90][91][100,101], circulating tumor DNA (ctDNA), genes [92][102], peptides [93][103], and blood plasma [94][104], are studied using SERS for disease identification. SERS tags that specifically bind to the targets under study are widely used for analyzing cancer samples [95][96][97][98][99][105,106,107,108,109]. Machine learning algorithms are used to analyze complex patterns and recognize buried signals overcoming noise from undesirable constituents of cells and other bio-fluids.2.2. SARS-CoV-2 and Other Respiratory Diseases
With the onset of the pandemic and the fast-spreading variants, there was a need to rapidly identify, detect, and quarantine the infected population. Surveying the presence of antibodies in large populations, often called a serological survey, was important to access the percentage of population infected and to monitor community transmission [100][236]. The dominant existing technique for the identification of SARS-CoV was PCR, which relies on analyzing the genetic material of the virus [101][237]. However, the test is expensive, thus preventing wide usage and also is time consuming. The Raman spectrum of a whole organism, including viruses, is contributed to by the proteins, carbohydrates, and nucleic acids that make up the organism [102][238]. The expression of these building blocks is controlled by the genetic material of the organism, hence helping in the unique identification [103][239]. SERS has enabled trace, point-of-care (POC), sample-collection-friendly, rapid, flexible, and cost-effective covid detection alternatives with the use of diverse nanomaterials [10][104][105][106][10,240,241,242]. In addition, both portable and handheld systems have indeed enabled point-of-care testing based on Raman spectroscopy [107][108][56,243]. SERS has also been widely used for the detection of other respiratory zoonotic diseases, such as H1N1, H7N9, H3N2, and H5N1; and other coronaviruses, such as MERS-CoV [109][110][244,245]. Often, machine learning algorithms are used in combination to enable the identification of patterns that are not apparent to the human eye [111][112][246,247]. The availability of large data and the ease of collection have accelerated the potential of machine learning algorithms in identifying viruses and their variants with reliable accuracies for POC devices [113][114][248,249]. In addition to trace identification, SERS has also enabled quantification of viral load to access the severity of the infection [115][116][250,251]. SERS in combination with LDA has been used for the rapid (2 min) identification of respiratory viruses, including SARS-CoV-2, human adenovirus type 7, and H1N1, using label-free silver nanoparticles [117][252]. Fe3O4@Ag nanoparticles tagged with specific antibodies were used for the detection of adenovirus and influenza virus [118][253]. Eleven different respiratory pathogens were identified using SERS, with nanoparticles tagged with nucleic acids achieving remarkable LODs in the sub-picomolar range [119][254]. Gold nanoparticles functionalized with a specific enzyme were used for the detection of S protein expressed by the COVID-19 viruses with SERS-based sensing in water [120][255]. Trace S protein detection has also been performed with SERS substrates enabling both chemical and electromagnetic enhancement [121][256] and using DNA-aptamer-based substrates, achieving a 0.7 fg mL−1 LOD [122][257]. Influenza-infected cells were identified based on proteins, using SERS and PCA [123][258]. Influenza and covid viruses were detected in human nasal fluid and saliva, using SERS [124][259], and also in untreated saliva [125][260]. A portable breath analyzer for covid detection based on the presence of organic volatile compounds was developed, achieving a sensitivity greater than 95% with less than 5 min of detection time [53]. A lateral-flow-immunoassay-based SERS was proposed for the quantitative detection of SARS-CoV-2 [126][261]. Similar work was performed for the trace detection of SARS-CoV-2 antibodies and spike proteins [127][128][129][262,263,264]. Li et al. optimized the silver nanostructures to increase the LOD for SARS-CoV-2 detection [130][265]. In a unique study, Kim et al. studied the efficacy of the Oxford–AstraZeneca vaccine by using SERS studies on tear samples and achieved excellent reproducibility and LOD in the femtomolar regime [131][266]. Machine learning algorithms such as PCA and SVM were used for the classification of normal and SARS-CoV-2 saliva samples with SERS data, with an accuracy of 95% [132][267]. Different respiratory viruses and their variants were identified using a silver-nanorods-based SERS sensor [133][268]. Different respiratory syncytial viruses have been identified and classified using SERS and classification algorithms such as PCA and HCA [134][269]. A deep-learning-based on-site SERS detection was developed to detect the SARS-CoV-2 virus based on the spike protein with 87% accuracy. This work also studied Raman modes of the spike protein theoretically and established a database [135][270]. Different variants of the SARS-CoV-2 virus, including wild-type, Alpha, Delta, and Omicron, were successfully identified using specific antibody-tagged 3D porous Ag-based SERS substrates [136][271]. SERS has also shown the potential of simultaneous detection of influenza virus (H1N1), SARS-CoV-2, and respiratory syncytial virus by using magnetic-tags-based SERS substrates with extended studies in throat swabs [137][272]. Label-free SERS was performed on serum samples of patients after 4 to 16 days of testing positive for COVID-19, and chemometric techniques were used to find significant difference in the SERS spectral features [138][273].3. SERS-Based Detection of Microorganisms
3.1. Bacteria Sensing
A bacterium is a living cell and falls under the class of prokaryotic microorganisms. Bacteria come in different shapes, including spheres, rods, spiral, and comma, and have a typical size of few micrometers [139][274]. Bacterial cells are omnipresent, as they are found in water, food, soil, air, and the human body, and, interestingly, the human body contains 10 times more bacterial cells than human cells. However, only 3% of the bacteria are pathogenic, while the other 97% are essential for the survival of different life forms on the earth [140][275]. The identification of bacteria is important to assess the quality and contamination of food, soil, and water as a measure of public health. In some cases, the presence of bacteria is also desirable to ensure the decomposition of undesirable contaminants through a process called bioremediation [141][142][143][276,277,278]. Conventionally, PCR, plate culture, and flow cytometry are used for the detection of bacteria. However, all of them are time-consuming and need 2 to 3 days to arrive at conclusions [144][279]. SERS-based sensing for bacteria is extensively used for its proven advantages of being specific, sensitive [145][146][147][280,281,282], rapid [148][283], and water compatible to perform in situ measurements [149][284], as well as having the ability to quantify [150][151][152][285,286,287] and potential for trace detection [153][154][155][156][157][158][288,289,290,291,292,293]. Point-of-care devices for detection of bacteria can also be realized through SERS [159][160][294,295]. The sensitivity of SERS even enabled the detection of the single bacterium [160][295]. It is even possible to distinguish between live and dead bacteria cells by using SERS [161][296]. With the use of appropriate machine learning techniques, researchers achieved strain-level distinction using SERS spectra [162][297]. A SERS biosensor using aptamer (aptamer–Fe3O4@Au) and antibiotic (Vancomycin–Au@MBA) molecules has been used for the detection and quantification of pathogenic bacteria achieving a LOD of 3 cells/mL [163][298]. Vancomycin tagged NPs were also used in fabricating a sandwich such as SERS substrate for identification and photothermal elimination of bacteria in blood samples [164][299]. Different bacteria species such as S. typhi, E. coli, and L. mono were identified using SERS with Fe3O4@Au magnetic nanoparticles and demonstrated good accuracy in real world samples such as beef, saliva, and urine [165][300]. Wang et al. have also used magnetic nanoparticles for the detection of S. aureus [166][167][301,302]. Inspired by polyphenolic chemistry, SERS substrates with metal phenolic networks were designed for the detection of E. coli and S. aureus [168][303]. In addition to E. coli detection, antibiotic susceptibility was studied using core–shell Au@Ag nanorods. This study was also extended to mice blood, implying practical usage [169][304]. Bacteria present in serum and human blood sampl was identified using SERS based sensing [170][171][305,306]. Polymer mats prepared by force spinning were used for the detection of S. aureus, P. aeruginosa, and S. Typhimurium in blood plasma [172][307]. Using external magnetic field and plasmonic magnetic nanoparticles, the sensitive detection of Gram-negative bacteria was performed by concentrating the sample to a small area [173][308]. Similar work was accomplished using a microfluidic device to analyze drinking water for bacterial contamination [174][309]. The quantification of Salmonella typhimurium was performed using 3D DNA-based SERS substrates [175][310]. SERS-based immunoassay was used for the ultrasensitive and quantitative detection of different bacteria species simultaneously [176][311]. Multiplexing was also demonstrated by Hayleigh et al. [177][312] and Gracie et al., who then went on to conduct quantification in multiplexing [178][313]. A ceramic-filter-based SERS substrate, along with metal nanoparticles, was used for the detection of E. coli and Shewanella putrefaciens [179][314]. Nine different species of E. coli were studied using a SERS microfluidic device and discriminated with 92% accuracy, using support vector machine analysis [180][315]. The label-free and portable detection of various foodborne bacteria was studied using SERS and different chemometric techniques, e.g., PCA and PLS-DA [181][316]. Silver nanoparticles synthesized using leaf extract were used for the detection of two bacteria species [182][317]. SERS, in combination with deep learning techniques, was used for the accurate identification of Staphylococcus aureus to achieve an accuracy of ~98% [183][318].3.2. Sensing of Biohazardous Molecules for Homeland Security
Bioterrorism is the new threat facing the world and is equally potent to cause large-scale destruction of civil, animal, and plant life. Often, biological agents are easy to prepare and scale up; can be contaminated in food, water, and soil; and are easy to carry, making them the future weapons. Many countries keep them in their military stockpiles despite the regulations [184][319]. According to the Centre for Disease Control and Prevention (CDC), a biohazardous material is defined as any infectious agent or biological material that poses a threat to human health, the environment, and animals. A review by Lister et al. summarized different biological agents that concern homeland security [185][320]. Different pathogens and biological agents, such as toxins, venom, and allergens, are some examples of biohazardous materials. Nerve agents are a big concern owing to their high solubility, high toxicity, and durability, with the Tokyo event in 1995 being an example [186][187][321,322]. Nerve agents can be classified into G-series, representing agents developed by Germans; V-series for venomous agents; GV series for the combination of G- and V-series; and Novichock series [188][323]. It is imperative to have a detection system that is sensitive, rapid, portable, and functional for different background media, such as liquids and gases, for the detection of these nerve agents. Plasmonic sensors are widely used for the detection of chemical and biological war threats [189][190][324,325]. Of all, SERS has its own advantages for the reasons discussed in the Introduction section and hence is widely used for the detection of biological threats, with a potential for field applications using portable devices [191][326] and chemometrics [192][327]. A sensitive and selective identification of the nerve agents Tabun, Cyclosarin, and VX was performed using gold- and silver-coated Si nanostructures both without [193][328] and with a tag (antidote) [194][329] in two different studies. VX and its hydrolysis products were studied elsewhere, too [195][196][330,331]. Sarin, an organophosphorus nerve agent, was detected using plasmonic Si nanocone structures [197][332]. Three nerve agents, i.e., isopropyl methylphosphonofluoridate (GB), pinacolyl methylphosphonofluoridate (GD), and cyclohexyl methylphosphonofluoridate (GF), were identified, and their hydrolysis degradation was distinguished using SERS [187][322]. A mustard simulant, pathogenic bacteria, and cyanide were detected using SERS [198][333]. A reproducible (7%), rapid (30 s), and sensitive (1 ppb) was used for the detection of a nerve simulant, pinacolyl methyl phosphonic acid (PMPA) [199][334]. Gaseous warfare agents such as dimethyl methylphosphonate were identified using SERS on LiCl microlenses [200][335]. Various G-series and VX nerve agents were identified using novel pinhole shell-isolated Au nanoparticles substrates achieving sensitivity of 10 ng/L and 20 ng/L, respectively [201][336]. Using plasmonic 3D fractal structures, a G-series nerve agent called dimethyl methylphosphonate (DMMP) was detected in the gaseous state, with a sensitivity of 12 ppmV [202][337]. Bacillus anthracis is a highly infectious bacteria that causes the fatal disease anthrax in humans. It is a cause for concern because of its recent usage as a biowarfare agent by many countries [203][338]. Farrell et al. summarized different anthrax biomarkers and existing detection techniques [204][339]. Plasmonic metal decorated anisotropic Ni nanostructures were used for detection of dipicolinic acid (DPA), a biomarker for anthrax [205][340]. Specifically, tagged SERS substrates were used for the detection of anthrax protective antigens, achieving a remarkable LOD of 1 pg/mL [206][341]. A magnetic microfluidic SERS sensor using specifically tagged Au nanoparticles was used for the detection of the anthrax biomarker poly-γ-D-glutamic acid, with an LOD of 100 pg/mL [207][342]. Reusable and sensitive laser-ablated Au nanostructures were used for the detection of dipicolinic acid (DPA) with a LOD of 0.83 pg/L and signal enhancement of ~1012 [208][343]. A selective SERS substrate that can discriminate between different strains of bacteria by specifically binding to Bacillus anthracis was designed with DPA as a biomarker [209][344]. Gold nanorods were also employed for the sensitive detection of DPA and anthrax-protective antigen [210][211][345,346]. The trace detection of DPA, equivalent to nearly 18 spores, was achieved using super-hydrophobic SERS sensors [212][347]. The effects of aggregation of NPs and pH on the SERS performance for the detection of components of cell wall and endospores of Bacillus thuringiensis were studied extensively [213][348]. Different chemical and biological warfare agents were classified using techniques such as PCA, PLS-DA, as well as hierarchical classification techniques based on the SERS spectra [193][214][328,349].4. Machine Learning in SERS-Based Biosensing
4.1. Introduction to Machine Learning
In recent times, machine learning is widely being used for many applications including spectroscopy for both data pre- and postprocessing. Machine learning (ML), as the name suggests, is a technique in which the algorithm learns patterns from the existing data and will attempt to make accurate predictions on the unknown based on the trained data. The potential for its ability to find complex patterns from big data sets has given an opportunity to extract and model data purposefully. There are different existing algorithms, both supervised and unsupervised, depending on the problem at hand. Deep learning is a subdomain of machine learning inspired by the human brain that uses multilayered neural networks for modeling data. Throughout this article, machine learning also implies deep learning techniques. Advances in computation facilities and with increasing availability and complexity of big data, deep learning, which is a kind of machine learning, has found its place. Some popular and relevant examples of ML being classification of emails as span and not span, identifying cancer in early stage using medical images, face recognition and weather prediction. ML algorithms can be broadly classified into three types, namely supervised for labeled observations, unsupervised for unlabeled observations, and reinforcement learning for models that learn from the errors to improve accuracy [215][350], as summarized in the Figure 2 below.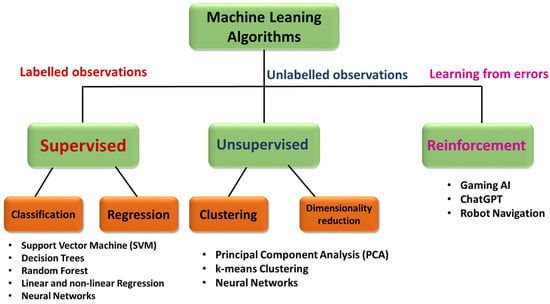
Figure 2. Flow chart illustrating the classification of different machine learning algorithms as supervised, unsupervised, and reinforcement models.
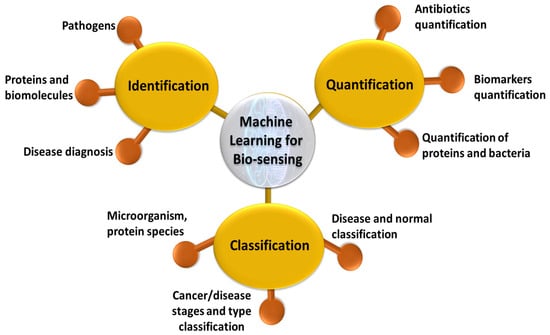
Figure 3.
Schematic of applications of machine learning for biosensing using SERS based plasmonic sensors.
4.2. Identification
SERS provides the vibrational fingerprint of many biomolecules, including amino acids, peptides, carbohydrates, pathogens, and nuclei acids [242][377]. It is also label free and non-destructive, making it desirable for in situ and rapid identification. Often in real-world situations of biology sample analysis, there are undesirable effects from background cell signals or with the similarity of spectra from two subspecies. Machine learning models can be successfully trained to capture these complex differences and distinguish two similar spectra devoid of the background helping in identification of the sample. CNNs were used for identification of cancer using SERS with gold multi-branched nanoparticles (AuMs), functionalized with different chemical groups, and achieved 100% accuracy in identifying the structural changes [243][378]. Drug-sensitive and drug-resistant bacterial strains were identified using SERS with a combination of CNNs and achieved 100% accuracy [244][379]. Different classification algorithms such as LDA, SVM, and KNN were used for the classification of bacterial extracellular vesicles for E. coli by strain and culture time using label-free approach of SERS [245][380]. SVM was successfully used for the identification of different drugs in human urine at trace levels with an accuracy greater than 92% [246][381]. A SERS chip was designed to identify a cancer marker, TIMP-1, and combined it with ML to identify lung and colon cancer in patients [247][382]. A label-free SERS, in combination with different machine learning algorithms, such as random forest, PCA-LDA, and decision trees, was used for the identification of colon cancer using serum samples. It was found that the random forest model outperformed the other two models in terms of accuracy and specificity [248][383]. SERS combined with ANN was used for the identification of different pollen samples despite many spectral contributions using Au NPs [249][384]. A microfluidic-chip-based SERS substrate with Au nanoparticles was used for the identification of l Jurkat, THP-1, and MONO-MAC-6 leukemia cell lysates, using SVM, and achieved 99% accuracy [250][385]. A lab-on-chip SERS device was fabricated and used for the successful identification of different species of mycobacteria [251][386]. The machine learning models PLS-DA and CNN were used to identify different stages of kidney malfunction in dialysis patients by using serum analysis by SERS. The CNN model achieved an accuracy of 96%, which is better than that of PLS-DA, with 84% [252][387]. The SVM outperformed other techniques in the identification of cyanobacteria, using SERS spectra of mutant and wild-type strains [253][388]. Using a dimensionality reduction technique, followed by a probabilistic ML model, SARS-CoV-2 identification was performed with an accuracy of ~85% [254][389]. SERS coupled with SVM was also used for the identification of lung cancers [86][96].4.3. Quantification
