Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Sirius Huang and Version 1 by Panagiotis ANTONIOU.
Beta-like globin gene expression is developmentally regulated during life by transcription factors, chromatin looping and epigenome modifications of the β-globin locus. Epigenome modifications, such as histone methylation/demethylation and acetylation/deacetylation and DNA methylation, are associated with up- or down-regulation of gene expression. The understanding of these mechanisms and their outcome in gene expression has paved the way to the development of new therapeutic strategies for treating various diseases, such as β-hemoglobinopathies.
- globin gene regulation
- epigenome editing
- β-hemoglobinopathies
1. Introduction
Epigenetic modifications, which are defined as chromatin alterations that result in a determined expression pattern without changing the genetic sequence, have been shown to occur during the fetal-to-adult hemoglobin switching (Figure 1). These potentially heritable modifications, which include DNA methylation and histone modifications (acetylation and methylation), can be cell-, tissue- and developmental-stage specific. Epigenetic modifications change the chromatin structure and DNA accessibility, and consequently affect gene expression, as epigenetic modifiers co-operate with TFs to repress or activate gene expression.
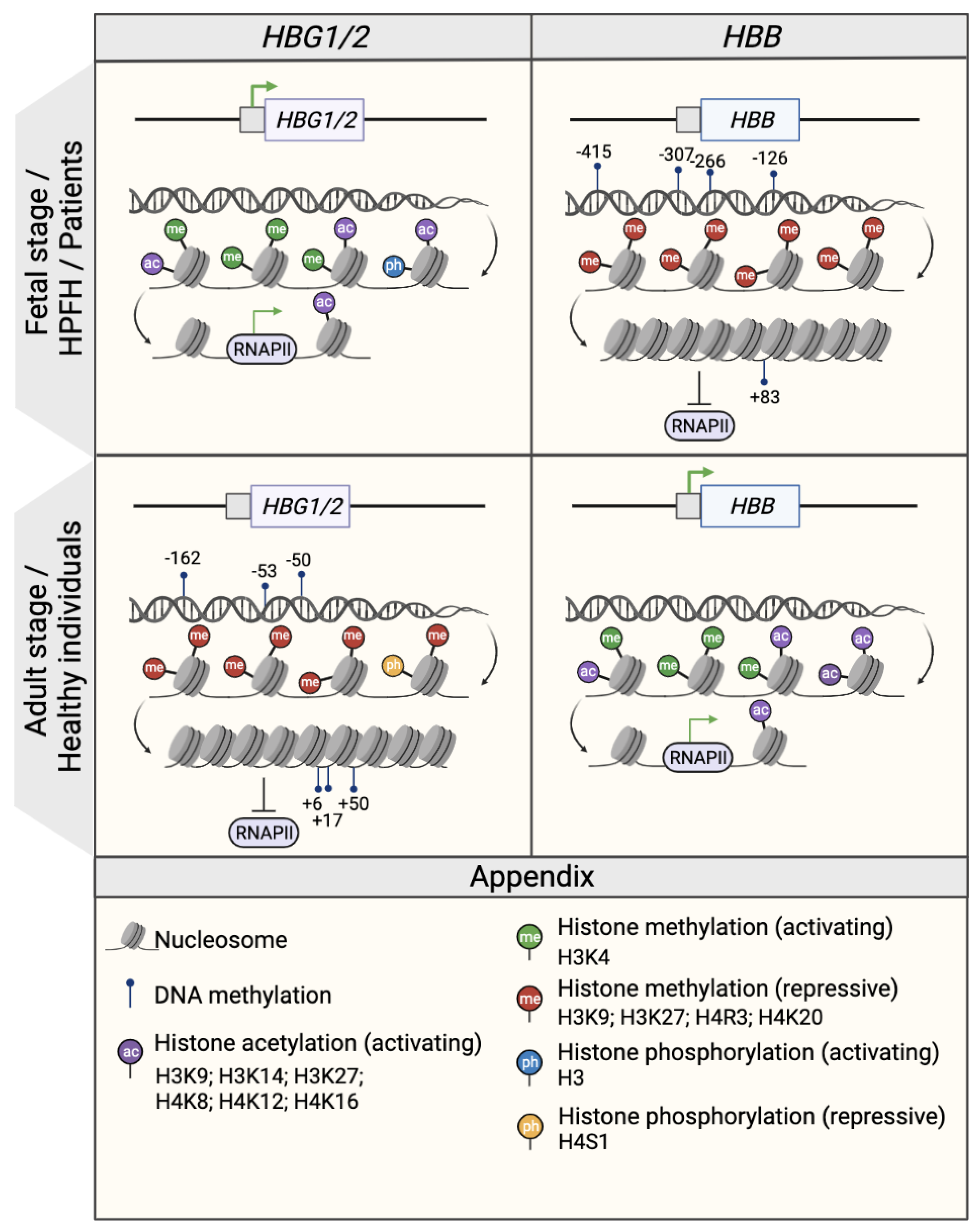
Figure 1. Epigenetic regulation of the β- and γ- globin gene expression. Schematic representation of the epigenetic profile of HBG1/2 and HBB promoters (represented by squares) in the fetal or fetal-like stages (fetal life, individuals with HPFH mutations and β-hemoglobinopathy patients; upper panel) and in the adult stage (healthy individuals; middle panel). The presence of DNA methylation and histone modifications (activating acetylation marks and activating or repressive methylation marks) is associated with an open or closed chromatin conformation that influences the accessibility of RNA polymerase II (RNAPII) and, consequently, the transcription rate as a whole. Green arrows indicate active transcription. CpG sites that are specifically methylated in the HBG1/2 genes in the adult stage are indicated and named after their distance from the transcription start site (e.g., −162, −53, −50, +6, +17, and +50). Created with BioRender.com.
2. DNA Methylation
The promoters of many human genes (including globin genes) contain Cytosine-phosphate Guanine (CpG) sites, which occur at a high frequency in regions known as “CpG islands”, and are amenable to epigenetic modifications. DNA methyltransferases (DNMT) deposit methyl groups in the fifth carbon atom of the cytosine of a CpG site. DNMT3A and DNMT3B mediate the de novo deposition of methyl groups in a non-methylated 5′-CpG-3′ and symmetrically to the non-methylated 3′-GpC-5′ of the complementary strand [49][1]. DNMT1 is responsible for the maintenance of the methylation of asymmetrically semi-methylated CpG sites upon DNA replication [50][2]. The ten-eleven translocation methyl-cytosine dioxygenase (TET) proteins TET1, TET2 and TET3 are responsible for DNA demethylation [51,52][3][4]. TET is responsible for the removal of the methyl group from a 5-methyl-Cytosine (5-mC) through the generation of a 5-hydroxy-methyl-cytosine (5-hmC) intermediate [51,52][3][4]. High DNA methylation levels normally characterize transcriptional inactive promoter regions, while hypomethylated promoters and promoters bearing the 5-hmC intermediate are generally highly expressed [53][5]. However, CpG methylation is not always associated with gene repression, such as when it occurs at enhancer regions. For this reason, some studies suggest that CpG methylation denotes active enhancer regions and might be required for depositing the active H3K27 histone acetylation mark (see below) [54][6].
In the γ-globin promoters, CpG sites span a 500 nt region centered on the transcription start site (−162, −53, −50, +6, +17, +50, CpG sites) [55,56,57][7][8][9] (Figure 31). Studies in erythroid cells obtained from human hematopoietic stem and progenitor cells (HSPCs) demonstrated that the γ-globin promoters were significantly hypomethylated in cord blood and fetal liver samples compared to adult bone marrow samples [55,56,57][7][8][9]. Conversely, the DNA methylation levels of the adult β-globin promoter were higher in fetal cells than in adult samples [55][7]. Moreover, an abnormal DNA hypomethylation pattern in the γ-globin promoters was associated with an increased HbF expression in β-thalassemia patient cells as compared to healthy donors [56][8] and in individuals with HPFH mutations [58][10]. By contrast, in a non-human primate baboon model, 5hmC levels positively correlated with γ-globin expression, although the 5hmC levels were substantially lower than 5Mc [53][5]. Interestingly, a missense mutation in the DNMT1 gene found in patients with HbE/β-thalassemia, leads to lower stability and enzymatic activity loss of DNMT1. This was associated with the reactivation of HbF due to the diminished recruitment of DNMT1 to the γ-globin promoters and the reduced methylation levels in CpG sites in the γ-globin promoters (−53, −50, +6, +17, and +50) of erythroid cells differentiated from patient HSPCs [59][11].
3. Histone Modifications
The DNA of eukaryotes is wrapped around histone octamers, i.e., nucleosomes, forming the chromatin. A 147 bp-long DNA fragment is wrapped around a histone octamer, composed of two copies of histone (H) 2A, H2B, H3 and H4, while histone H1 binds to the linker DNA between two nucleosomes. A variety of post-translational modifications (e.g., methylation, acetylation and phosphorylation) in different amino acid residues of the histones have been identified and associated with distinct regulatory functions [60,61,62,63][12][13][14][15].
3.1. Histone Methylations
The transfer of methyl groups to histone amino acid residues by methyl-transferase proteins (lysine methyltransferase (KMT) and protein arginine methyltransferase (PRMT)) is associated either with an open chromatin state and gene activation, or to a compact chromatin state and gene repression. For instance, the H3 lysine 4 monomethylation, dimethylation or trimethylation (H3K4me1, H3K4me2 and H3K4me3, respectively) and the asymmetric H4 arginine 3 dimethylation (H4R3me2a) mark transcriptionally active genomic regions (with H3K4me1 typically marking enhancers, H3K4me2 marking promoters and enhancers, and H3K4me3 marking promoters). On the other hand, the deposition of methyl, dimethyl or trimethyl groups on H3 lysine 9 or lysine 27 (H3K9me1, H3K9me2, H3K9me3, H3K27me2 and H3K27me3, respectively) or symmetric H4 arginine 3 (H4R3me2s) or H4 lysine 20 (H4K20me3) is associated with transcription inactivation [64][16]. Interestingly, the simultaneous presence of activating H3K4me3 and repressive H3K27me3 characterizes a bivalent chromatin state typical of genes poised for rapid activation [61][13]. These modifications can be removed by demethylases (e.g., lysine-specific histone demethylase 1 (KDM1A), also known as lysine-specific demethylase (LSD1)).
The role of histone methylation in the regulation of the β-globin genes has been studied for decades (Figure 31). The developmental-stage specific pattern of histone modifications along the β-globin locus was analyzed by comparing human embryonic cell-, fetal liver- and bone marrow-derived erythroblasts. The three sources have a distinct globin expression profile corresponding to an embryonic/fetal, fetal and adult state, respectively. Accordingly, the presence of the active H3K4me3 mark is correlated with gene activity in the different cell types [65][17]. Studies have revealed a correlation between the type of histone methylation and the activation or repression of globin genes [66,67,68][18][19][20]. For instance, in human adult erythroid cells, the activating H3K4me2/me3 and the repressive H3K9me1 marks are present at the β- and γ-globin promoters, respectively [43,66][18][21]. Interestingly, upon administration of tranylcypromine (TCP), a KDM1A inhibitor [69][22], the activating H3K4me2 mark increased in the γ-globin but not in the β-globin promoter along differentiation, suggesting that KDM1A is responsible for the repression of the fetal genes by removing the activating H3K4me2 mark.
Additional proteins are involved in the determination of the histone methylation status of the β-globin locus. In particular, the expression of EHMT1/2 as well as the global H3K9me2 levels decrease along the erythroid differentiation in cells derived from adult HSPCs [70][23]. Pharmacological inhibition of EHMT1/2 (using UNC0638) or shRNA-mediated knockdown of EHMT1/2 led to decreased levels of H3K9me2 and, consequently, HbF reactivation [70,71][23][24]. However, the reactivation of HbF observed upon EHMT1/2 downregulation was associated with an additional activating event, the acetylation of H3 lysine 9 (H3K9Ac) at the HBG1/2 promoters, which is typically associated with gene activation (see paragraph “Histone acetylations”) [70][23]. Promisingly, UNC0638 also led to high levels of HbF expression in erythroid cells from β-thalassemia patients [72][25]. FTX-6058, and inhibitor of EDD, was also tested in healthy volunteers and induced high HbF levels without major side effects (NCT04586985).
Methylation of arginine 3 on H4 also plays an important role in globin gene regulation. In human erythroid cell lines, PRMT5 induces symmetric H4R3me2 in the γ-globin promoters [73][26]. The developmental-stage specific pattern of H4R3me2s marks along the β-globin locus was observed in primary erythroid cells. H4R3me2s levels in the γ-globin promoters were higher in human bone marrow-derived erythroid cells compared to cord blood-derived erythroid cells [73][26]. Conversely, H4R3me2s marks and PRMT5 were absent from the β-globin promoters of human bone marrow-derived erythroid cells. Small-molecule inhibition of PRMT5 (using adenosine-2′,3′-dialdehyde (Adox)) in erythroid cells derived from adult bone marrow HSPCs reduce H4R3me2s levels at the γ-globin promoters with a concomitant HbF reactivation. PRMT5 is thought to be recruited at the γ-globin promoters by LYAR. In fact, upon LYAR knockdown, both PRMT5 occupancy and H4R3me2 levels decrease at the γ- globin promoters [74][27].
An interplay between the different histone methylations and between histone methylations and DNA methylation has been reported at the β-like globin promoters. In human erythroid cell lines, PRMT5-induced symmetric H4R3me2 in the γ-globin promoters is associated with the recruitment of DNMT3A that methylates close CpG sites, highlighting the interplay between histone and DNA methylation [73][26]. Furthermore, in human erythroid cell lines, PRMT5 also induces the deposition of additional repressive histone methylation marks in the γ-globin promoters, such as H4K20me3, H3K9me3 and H3K27me3, likely by recruiting other repressor complexes such as KMT5B (also known as SUV4-20h1), a lysine methyltransferase that deposits the repressive H4K20me3 mark. The H4K20me3 mark was higher at the embryonic/fetal ε- and γ-globin promoters in human bone marrow-derived erythroblasts as compared to cord blood-derived erythroid cells. Accordingly, shRNA-mediated downregulation of SUV4-20h1 led to γ-globin gene reactivation in adult cells [64][16].
3.2. Histone Acetylations
The deposition of acetyl groups on histone lysine residues loosens the interactions between histones and DNA due to a neutralization of the positive histone charge [75,76][28][29]. As a result, chromatin is in an open state, and TFs can bind to the DNA resulting in transcription activation. The proteins that are responsible for histone acetylation are histone acetyl-transferases (HAT; for example, p300), which are lysine acetyl-transferases (KAT) that deposit acetyl groups in lysine residues of the histone tail. Conversely, lysine deacetylases (KDAC), including histone deacetylases (HDAC), can remove the acetyl groups from histones, thus compacting the chromatin. There are four classes of HDACs, Rpd3-like proteins (class I; HDAC1, HDAC2, HDAC3 and HDAC8), Hda1-like proteins (class II; HDAC4, HDAC5, HDAC6, HDAC7, HDAC9 and HDAC10), HDAC11 (class IV) and Sir2-like proteins (class III; SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6 and SIRT7).
Histone acetylation plays a dynamic role in the developmental-stage specific regulation of β-like globin gene expression (Figure 31). A positive correlation has been established between the β-like globin gene expression levels and H3 and H4 acetylation in the LCR and in the β-like globin genes in murine fetal liver cells and erythroid cell lines harboring the human β-globin locus [77,78,79][30][31][32].
Studies using human fetal and adult erythroid cells have shown that the levels of H3 acetylation in the LCR are stable during development, while the same marker in the β-globin coding regions positively correlates with HBB expression levels [65,80][17][33]. In general, in human primary cells, when a β-like globin gene is activated (e.g., the γ-globin gene in fetal erythroblasts), there is an H3 loss from the promoter and a strong acetylation of the remaining histones leading to a more open chromatin state [80][33]. In fact, human primitive-like, fetal and adult erythroid cells display H3 lysine 9 or 27 hyperacetylation in the ε-, γ- and β-globin gene promoters, respectively [55,66,67,80][7][18][19][33]
Interestingly, histone deacetylase activity is not always correlated with γ-globin repression. In fact, SIRT1, an histone deacetylase has been positively associated with increased γ-globin expression, and SIRT1 activators enhance the expression of γ-globin in primary human erythroid cells [81][34]. In fetal cord blood erythroid cells, the levels of SIRT1 are higher as compared to adult bone marrow cells [81][34]. SIRT1 was found to enhance the looping between the LCR and the γ-globin promoters, and this was also associated with an indirect increase in the levels of the activating H4K16ac mark in the γ-globin promoters [81][34]. Therefore, SIRT1 activators could potentially be used for reactivating HbF in hemoglobinopathy patients.
The interplay between different types of histone modifications also characterizes the β-globin locus regulation and has also been investigated at the level of enhancers (typically marked by H3K4me1) in human erythroid cell lines. In the β-globin LCR, H3K4me1 was correlated with histone acetylation, namely, H3K27ac. Interestingly, the loss of H3K4me1 led to a concomitant reduction in H3K27ac, but not vice versa, indicating a hierarchy that places methylation upstream of acetylation in the β-globin locus regulation [82][35].
Histone acetylations also cooperate with DNA methylation in regulating globin gene expression. In human cells, H3 and H4 acetylation regulates the β-globin genes in concert with other histone modifications (e.g., histone methylations and phosphorylation) and DNA methylation. Along the human erythroid differentiation, the activating H3K9ac marks in the β-globin locus overlap with histone methylation marks (H3K4me1 and H3K4me3), and 5-hmC [83][36], highlighting the interplay between different epigenetic marks to control gene expression. Furthermore, PRMT5 [64][16] or LYAR [74][27] downregulation in human erythroid cell lines led not only to the eradication of the H4R3me2s repressive mark, but also led to a reduction in the repressive H4 serine 1 phosphorylation (H4S1ph) mark and an increase in the activating H4K12ac, H4K8ac and H3K9ac marks. Inhibition of PRMT5 by Adox in adult bone marrow erythroid cells also increased the H4 acetylation levels at the γ-globin promoters [84][37].
4. The Interplay between Epigenetic Modifiers, Transcription Factors and Chromatin Looping
The fine-tuning of globin gene expression is mediated by the direct or indirect repressive or activating effects of TFs that bind to the β-globin locus, and by an interplay between these TFs and DNA or histone modifiers that alter the epigenetic profile of the locus and, thus, its chromatin accessibility. By way of example, the GATA1 TF interacts with the CBP/p300 acetyltransferase during erythropoiesis [85][38]. With regard to the fetal-to-adult Hb switching, the major players of this interplay are the BCL11A and LRF TFs that interact with co-factors including epigenome modifiers. BCL11A and LRF interact with the NuRD complex, which contains HDACs and MBD2. MBD2 binds to methylated CpG regions and recruits other NuRD components to repress gene expression [86][39]. BCL11A also interacts with histone demethylases, such as KDM1A, which removes activating histone methylation marks in the γ-globin promoters [43][21]. Another repressive complex that regulates the expression of γ-globin genes is the direct repeat erythroid-definitive (DRED) complex that contains KDM1A, DNMT1 and testicular receptor 2 and 4 (TR2/TR4)TF [43,87][21][40].
Finally, the specific binding of TFs to the β-like globin genes and the chromatin remodeling through LCR-mediated looping formation is required for the regulated expression of β-like globin genes. The chromatin looping drives the γ-globin or β-globin gene expression in fetal and adult erythroblasts, respectively. The looping is mediated by a protein complex containing LDB1, LMO2, GATA1, TAL1 and E2A and facilitates the juxtaposition of the LCR to the γ- or the β-globin gene promoter, thus promoting their expression [32,33,34,35,36][41][42][43][44][45]. The recruitment of this protein complex inducing chromatin looping is also associated with epigenetic changes in the β-globin locus. In fact, in a murine erythroid cell line, GATA1 induces LDB1-mediated chromatin looping and H3K4me3 deposition at the β-globin gene to regulate its expression [88][46]. Furthermore, in a human fetal erythroid cell line, the ETO2 transcriptional co-repressor impairs the binding of this protein complex to the β-globin locus and, as a consequence, the looping between the LCR to the γ-globin gene. This is paralleled by the recruitment of the NuRD complex, the decrease in H3K27ac and H3K9ac at the β-globin locus, and the repression of the γ-globin genes [89][47]. Taken together, these results indicate a tight interplay between chromatin looping and epigenetic modifications at the β-globin locus to promote proper β-like globin gene regulation.
References
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell 1999, 99, 247–257.
- Li, E.; Bestor, T.H.; Jaenisch, R. Targeted Mutation of the DNA Methyltransferase Gene Results in Embryonic Lethality. Cell 1992, 69, 915–926.
- He, Y.-F.; Li, B.-Z.; Li, Z.; Liu, P.; Wang, Y.; Tang, Q.; Ding, J.; Jia, Y.; Chen, Z.; Li, L.; et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science 2011, 333, 1303–1307.
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science 2011, 333, 1300–1303.
- Ruiz, M.A.; Rivers, A.; Ibanez, V.; Vaitkus, K.; Mahmud, N.; DeSimone, J.; Lavelle, D. Hydroxymethylcytosine and Demethylation of the γ-Globin Gene Promoter during Erythroid Differentiation. Epigenetics 2015, 10, 397–407.
- Charlet, J.; Duymich, C.E.; Lay, F.D.; Mundbjerg, K.; Dalsgaard Sørensen, K.; Liang, G.; Jones, P.A. Bivalent Regions of Cytosine Methylation and H3K27 Acetylation Suggest an Active Role for DNA Methylation at Enhancers. Mol. Cell 2016, 62, 422–431.
- Mabaera, R.; Richardson, C.A.; Johnson, K.; Hsu, M.; Fiering, S.; Lowrey, C.H. Developmental- and Differentiation-Specific Patterns of Human Gamma- and Beta-Globin Promoter DNA Methylation. Blood 2007, 110, 1343–1352.
- Bao, X.; Zuo, Y.; Chen, D.; Zhao, C. DNA Methylation Patterns of β-Globin Cluster in β-Thalassemia Patients. Clin. Epigenetics 2020, 12, 187.
- Lessard, S.; Beaudoin, M.; Benkirane, K.; Lettre, G. Comparison of DNA Methylation Profiles in Human Fetal and Adult Red Blood Cell Progenitors. Genome. Med. 2015, 7, 1.
- Goren, A.; Simchen, G.; Fibach, E.; Szabo, P.E.; Tanimoto, K.; Chakalova, L.; Pfeifer, G.P.; Fraser, P.J.; Engel, J.D.; Cedar, H. Fine Tuning of Globin Gene Expression by DNA Methylation. PLoS ONE 2006, 1, e46.
- Gong, Y.; Zhang, X.; Zhang, Q.; Zhang, Y.; Ye, Y.; Yu, W.; Shao, C.; Yan, T.; Huang, J.; Zhong, J.; et al. A Natural DNMT1 Mutation Elevates the Fetal Hemoglobin Level via Epigenetic Derepression of the γ-Globin Gene in β-Thalassemia. Blood 2021, 137, 1652–1657.
- Tan, M.; Luo, H.; Lee, S.; Jin, F.; Yang, J.S.; Montellier, E.; Buchou, T.; Cheng, Z.; Rousseaux, S.; Rajagopal, N.; et al. Identification of 67 Histone Marks and Histone Lysine Crotonylation as a New Type of Histone Modification. Cell 2011, 146, 1016–1028.
- Zhou, V.W.; Goren, A.; Bernstein, B.E. Charting Histone Modifications and the Functional Organization of Mammalian Genomes. Nat. Rev. Genet. 2011, 12, 7–18.
- Romano, O.; Petiti, L.; Felix, T.; Meneghini, V.; Portafax, M.; Antoniani, C.; Amendola, M.; Bicciato, S.; Peano, C.; Miccio, A. GATA Factor-Mediated Gene Regulation in Human Erythropoiesis. iScience 2020, 23, 101018.
- Allfrey, V.G.; Faulkner, R.; Mirsky, A.E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. USA 1964, 51, 786–794.
- Rank, G.; Cerruti, L.; Simpson, R.J.; Moritz, R.L.; Jane, S.M.; Zhao, Q. Identification of a PRMT5-Dependent Repressor Complex Linked to Silencing of Human Fetal Globin Gene Expression. Blood 2010, 116, 1585–1592.
- Chang, K.-H.; Fang, X.; Wang, H.; Huang, A.; Cao, H.; Yang, Y.; Bonig, H.; Stamatoyannopoulos, J.A.; Papayannopoulou, T. Epigenetic Modifications and Chromosome Conformations of the Beta Globin Locus throughout Development. Stem. Cell Rev. Rep. 2013, 9, 397–407.
- Xu, J.; Shao, Z.; Glass, K.; Bauer, D.E.; Pinello, L.; Van Handel, B.; Hou, S.; Stamatoyannopoulos, J.A.; Mikkola, H.K.A.; Yuan, G.-C.; et al. Combinatorial Assembly of Developmental Stage-Specific Enhancers Controls Gene Expression Programs during Human Erythropoiesis. Dev. Cell 2012, 23, 796–811.
- Hsu, M.; Richardson, C.A.; Olivier, E.; Qiu, C.; Bouhassira, E.E.; Lowrey, C.H.; Fiering, S. Complex Developmental Patterns of Histone Modifications Associated with the Human Beta-Globin Switch in Primary Cells. Exp. Hematol. 2009, 37, 799–806.e4.
- Kim, A.; Kiefer, C.M.; Dean, A. Distinctive Signatures of Histone Methylation in Transcribed Coding and Noncoding Human Beta-Globin Sequences. Mol. Cell Biol. 2007, 27, 1271–1279.
- Xu, J.; Bauer, D.E.; Kerenyi, M.A.; Vo, T.D.; Hou, S.; Hsu, Y.-J.; Yao, H.; Trowbridge, J.J.; Mandel, G.; Orkin, S.H. Corepressor-Dependent Silencing of Fetal Hemoglobin Expression by BCL11A. Proc. Natl. Acad. Sci. USA 2013, 110, 6518–6523.
- Shi, L.; Cui, S.; Engel, J.D.; Tanabe, O. Lysine-Specific Demethylase 1 Is a Therapeutic Target for Fetal Hemoglobin Induction. Nat. Med. 2013, 19, 291–294.
- Renneville, A.; Van Galen, P.; Canver, M.C.; McConkey, M.; Krill-Burger, J.M.; Dorfman, D.M.; Holson, E.B.; Bernstein, B.E.; Orkin, S.H.; Bauer, D.E.; et al. EHMT1 and EHMT2 Inhibition Induces Fetal Hemoglobin Expression. Blood 2015, 126, 1930–1939.
- Krivega, I.; Byrnes, C.; de Vasconcellos, J.F.; Lee, Y.T.; Kaushal, M.; Dean, A.; Miller, J.L. Inhibition of G9a Methyltransferase Stimulates Fetal Hemoglobin Production by Facilitating LCR/γ-Globin Looping. Blood 2015, 126, 665–672.
- Nualkaew, T.; Khamphikham, P.; Pongpaksupasin, P.; Kaewsakulthong, W.; Songdej, D.; Paiboonsukwong, K.; Sripichai, O.; Engel, J.D.; Hongeng, S.; Fucharoen, S.; et al. UNC0638 Induces High Levels of Fetal Hemoglobin Expression in β-Thalassemia/HbE Erythroid Progenitor Cells. Ann. Hematol. 2020, 99, 2027–2036.
- Zhao, Q.; Rank, G.; Tan, Y.T.; Li, H.; Moritz, R.L.; Simpson, R.J.; Cerruti, L.; Curtis, D.J.; Patel, D.J.; Allis, C.D.; et al. PRMT5-Mediated Methylation of Histone H4R3 Recruits DNMT3A, Coupling Histone and DNA Methylation in Gene Silencing. Nat. Struct. Mol. Biol. 2009, 16, 304–311.
- Ju, J.; Wang, Y.; Liu, R.; Zhang, Y.; Xu, Z.; Wang, Y.; Wu, Y.; Liu, M.; Cerruti, L.; Zou, F.; et al. Human Fetal Globin Gene Expression Is Regulated by LYAR. Nucleic Acids Res. 2014, 42, 9740–9752.
- Shvedunova, M.; Akhtar, A. Modulation of Cellular Processes by Histone and Non-Histone Protein Acetylation. Nat. Rev. Mol. Cell Biol. 2022, 23, 329–349.
- López-Bañuelos, L.; Vega, L. Inhibition of Acetylation, Is It Enough to Fight Cancer? Crit. Rev. Oncol. Hematol. 2022, 176, 103752.
- Forsberg, E.C.; Downs, K.M.; Christensen, H.M.; Im, H.; Nuzzi, P.A.; Bresnick, E.H. Developmentally Dynamic Histone Acetylation Pattern of a Tissue-Specific Chromatin Domain. Proc. Natl. Acad. Sci. USA 2000, 97, 14494–14499.
- Im, H.; Grass, J.A.; Christensen, H.M.; Perkins, A.; Bresnick, E.H. Histone Deacetylase-Dependent Establishment and Maintenance of Broad Low-Level Histone Acetylation within a Tissue-Specific Chromatin Domain. Biochemistry 2002, 41, 15152–15160.
- Schübeler, D.; Francastel, C.; Cimbora, D.M.; Reik, A.; Martin, D.I.; Groudine, M. Nuclear Localization and Histone Acetylation: A Pathway for Chromatin Opening and Transcriptional Activation of the Human Beta-Globin Locus. Genes Dev. 2000, 14, 940–950.
- Yin, W.; Barkess, G.; Fang, X.; Xiang, P.; Cao, H.; Stamatoyannopoulos, G.; Li, Q. Histone Acetylation at the Human Beta-Globin Locus Changes with Developmental Age. Blood 2007, 110, 4101–4107.
- Dai, Y.; Chen, T.; Ijaz, H.; Cho, E.H.; Steinberg, M.H. SIRT1 Activates the Expression of Fetal Hemoglobin Genes. Am. J. Hematol. 2017, 92, 1177–1186.
- Kang, Y.; Kim, Y.W.; Kang, J.; Kim, A. Histone H3K4me1 and H3K27ac Play Roles in Nucleosome Eviction and ERNA Transcription, Respectively, at Enhancers. FASEB J. 2021, 35, e21781.
- Madzo, J.; Liu, H.; Rodriguez, A.; Vasanthakumar, A.; Sundaravel, S.; Caces, D.B.D.; Looney, T.J.; Zhang, L.; Lepore, J.B.; Macrae, T.; et al. Hydroxymethylation at Gene Regulatory Regions Directs Stem/Early Progenitor Cell Commitment during Erythropoiesis. Cell. Rep. 2014, 6, 231–244.
- He, Y.; Rank, G.; Zhang, M.; Ju, J.; Liu, R.; Xu, Z.; Brown, F.; Cerruti, L.; Ma, C.; Tan, R.; et al. Induction of Human Fetal Hemoglobin Expression by Adenosine-2’,3’-Dialdehyde. J. Transl. Med. 2013, 11, 14.
- Blobel, G.A.; Nakajima, T.; Eckner, R.; Montminy, M.; Orkin, S.H. CREB-Binding Protein Cooperates with Transcription Factor GATA-1 and Is Required for Erythroid Differentiation. Proc. Natl. Acad. Sci. USA 1998, 95, 2061–2066.
- Desai, M.A.; Webb, H.D.; Sinanan, L.M.; Scarsdale, J.N.; Walavalkar, N.M.; Ginder, G.D.; Williams, D.C. An Intrinsically Disordered Region of Methyl-CpG Binding Domain Protein 2 (MBD2) Recruits the Histone Deacetylase Core of the NuRD Complex. Nucleic Acids Res. 2015, 43, 3100–3113.
- Cui, S.; Kolodziej, K.E.; Obara, N.; Amaral-Psarris, A.; Demmers, J.; Shi, L.; Engel, J.D.; Grosveld, F.; Strouboulis, J.; Tanabe, O. Nuclear Receptors TR2 and TR4 Recruit Multiple Epigenetic Transcriptional Corepressors That Associate Specifically with the Embryonic β-Type Globin Promoters in Differentiated Adult Erythroid Cells▿. Mol. Cell Biol. 2011, 31, 3298–3311.
- Wadman, I.A. The LIM-Only Protein Lmo2 Is a Bridging Molecule Assembling an Erythroid, DNA-Binding Complex Which Includes the TAL1, E47, GATA-1 and Ldb1/NLI Proteins. EMBO J. 1997, 16, 3145–3157.
- Song, S.-H.; Hou, C.; Dean, A. A Positive Role for NLI/Ldb1 in Long-Range β-Globin Locus Control Region Function. Mol. Cell 2007, 28, 810–822.
- Deng, W.; Rupon, J.W.; Krivega, I.; Breda, L.; Motta, I.; Jahn, K.S.; Reik, A.; Gregory, P.D.; Rivella, S.; Dean, A.; et al. Reactivation of Developmentally Silenced Globin Genes by Forced Chromatin Looping. Cell 2014, 158, 849–860.
- Krivega, I.; Dale, R.K.; Dean, A. Role of LDB1 in the Transition from Chromatin Looping to Transcription Activation. Genes Dev. 2014, 28, 1278–1290.
- Yun, W.J.; Kim, Y.W.; Kang, Y.; Lee, J.; Dean, A.; Kim, A. The Hematopoietic Regulator TAL1 Is Required for Chromatin Looping between the β-Globin LCR and Human γ-Globin Genes to Activate Transcription. Nucleic Acids Res. 2014, 42, 4283–4293.
- Deng, W.; Lee, J.; Wang, H.; Miller, J.; Reik, A.; Gregory, P.D.; Dean, A.; Blobel, G.A. Controlling Long-Range Genomic Interactions at a Native Locus by Targeted Tethering of a Looping Factor. Cell 2012, 149, 1233–1244.
- Guo, X.; Plank-Bazinet, J.; Krivega, I.; Dale, R.K.; Dean, A. Embryonic Erythropoiesis and Hemoglobin Switching Require Transcriptional Repressor ETO2 to Modulate Chromatin Organization. Nucleic Acids Res. 2020, 48, 10226–10240.
More
