Antibiotic resistance continues to evolve and spread beyond all boundaries, resulting in an increase in morbidity and mortality for non-curable infectious diseases. Due to the failure of conventional antimicrobial therapy and the lack of introduction of a novel class of antibiotics, novel strategies have emerged to combat these multidrug-resistant infectious microorganisms.
- multi drug resistance
- gram-negative
- bacteria
1. Introduction
-
By blocking the inactivating enzyme;
-
By tackling the mechanisms that impair the intracellular concentration required for antibacterial activity;
-
By bypassing target mutation.
2. Mechanism of Resistance
2.1. Membrane Impermeability
In Gram-negative bacteria, porins are transmembrane proteins facilitating the transport of hydrophilic molecules across the outer membrane, while hydrophobic molecules can diffuse through the phospholipid bilayer. Consequently, charged antibiotics, such as ß-lactams, will cross the outer membrane by way of the porin in order to reach the periplasmic target [8]. To limit the antibiotic action, the Gram-negative bacteria have developed mechanisms that decrease their membrane diffusion. Changes in outer membrane permeability impair the influx of hydrophilic antibiotics using porins like β-lactams and fluoroquinolones [9]. An alteration in the porin expression or in porin integrity leads to a decrease in internal concentration and therefore resistance to these antibiotics. For example, Klebsiella aerogenes (formerly Enterobacter) becomes resistant to cephalosporins because of an amino acid mutation located inside the eyelet of porin that strongly reduces the pore diameter. This structural mutation generates a restricted diffusion of antibiotics through porins, conferring this peculiar resistance [10].2.1.1. Antibiotic Target Modification
The antibiotic affinity on a target is a particularly important event during the fight against a bacterial infection. A target modification caused by spontaneous mutation at the gene level (substitutions of amino acids on binding sites located on the target surface), alters the affinity of the antibiotic, favoring the emergence of a resistance level [11]. The involvement of mutations in the QRDR region in a quinolone target, gyrase and topoisomerase enzymes, is extensively documented in Gram-negative pathogens [12].2.1.2. Antibiotic Inactivation
Numerous enzymes produced by bacteria can inactivate the antibiotic either by modifying or hydrolyzing it. Thus, these enzymes can act against β-lactams, aminoglycosides, chloramphenicol, or antibiotics belonging to the macrolide-lincosamide-streptogramin (MLS) family [13]. Inactivation of antibiotics occurs either by hydrolysis or by association with modifying metabolism or elimination (phosphorylation, acetylation, etc.). The β-lactam antibiotics exhibit a β-lactam ring which is the effective part of the molecules, as it inhibits the transpeptidase involved in the synthesis of the Gram-negative bacterial wall; the hydrolysis of this part is one of the degradation mechanisms involved in antibiotic resistance. Thus, the bacterium is able to produce enzymes, termed β-lactamases, able to hydrolyze the β-lactam ring 1 [14][15].2.1.3. Efflux Pumps
Efflux pumps are transmembrane systems by which the cells expel toxic compounds outside using active transport (energy-dependent), contributing to the bacterial resistance mechanism. A decrease in internal antimicrobial amount under the biological required concentration impairs and limits the antibiotic action [16]. The efflux pumps are classified into six families: the ABC (Adenosine triphosphate-Binding Cassette) family, the MFS (Major Facilitator Superfamily) family, the MATE (Multidrug And Toxic Compound Extrusion) family, the SMR (Small Multidrug Resistance) family, the RND family (Resistance Nodulation cell Division), and finally the PACE family (Proteobacterial Antimicrobial Compounds Efflux) [17].2.2. Strategies to Circumvent Gram-Negative Bacterial Resistance
One of the most promising approaches to circumvent Gram-negative bacterial resistance requires the combination of several types of molecules (e.g., enzyme inhibitor + antibiotic or membrane permeabilizer + antibiotic), for which the biological action can be either additive or synergistic. The additive effect occurs when the observed effect is the sum of the activity of the two partners, while the synergy corresponds to a final action greater than the sum of the activity of the two individual partners [18]. The use of this combination can also minimize the risk of the emergence/spreading of bacterial resistance for the treatment of recurrent infections [19].2.2.1. Drug Combination
To restore the activity of an antibiotic impaired by a resistance mechanism, an effective strategy consists in associating two antibiotics during patient treatment. The development of the hybrid antibiotic approach needs a chemical covalent connection between two molecules; generating a single hybrid drug that targets bacterial cells can mitigate bacterial resistance mechanisms. In this context, a combination of two antibiotics, such as a fluoroquinolone (ciprofloxacin) and an aminoglycoside (neomycin B), demonstrated a significant delay in resistance emergence compared to each component taken alone, and this is thanks to the multiple antibiotic resistance operon “Mar” involved in the regulation of efflux pump expression and OM permeability [20].
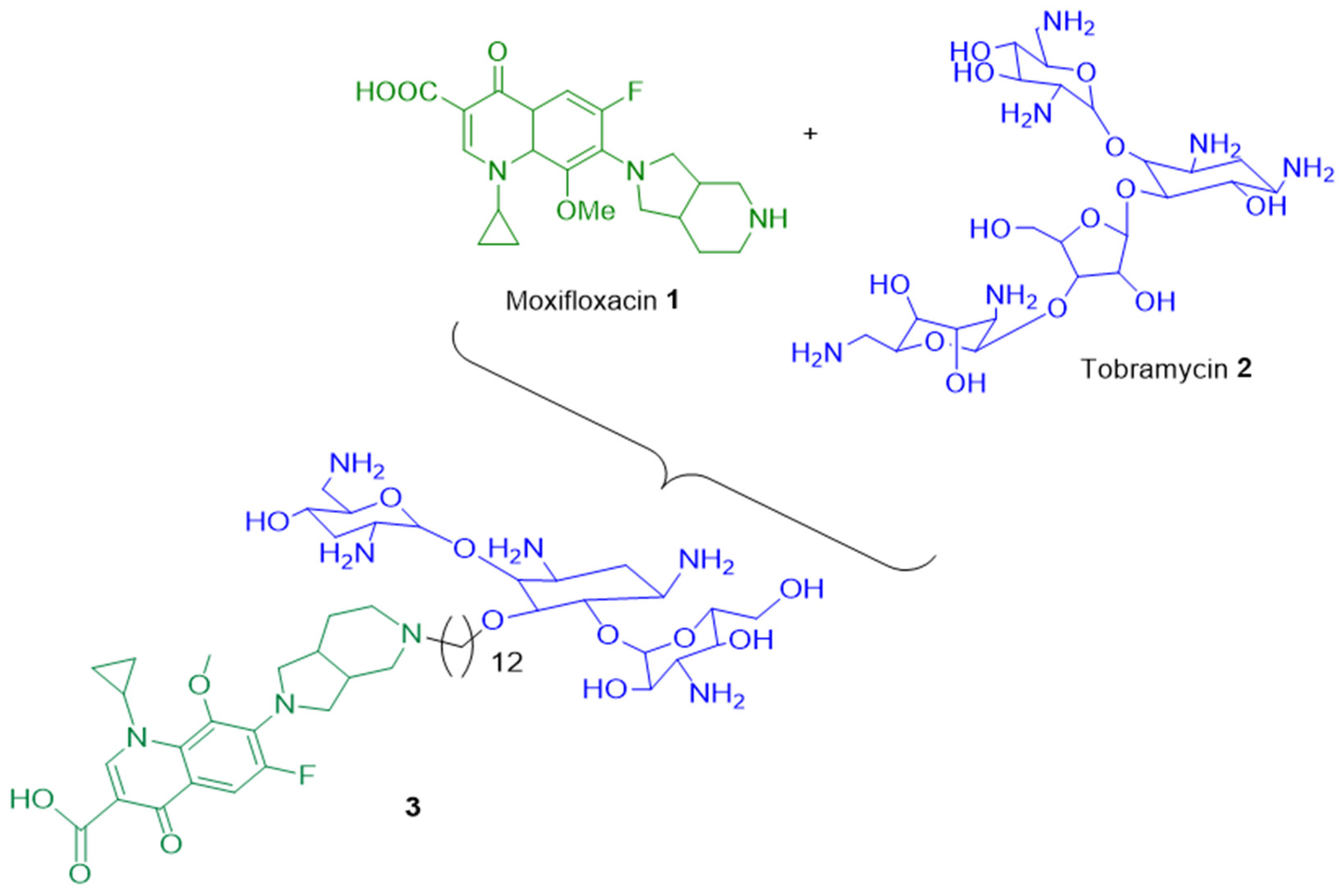
In the multiresistant (MDR) bacterium Pseudomonas aeruginosa, it has been shown that the hybrid tobramycin-moxifloxacin 3 led to better intracellular concentration of antibacterial molecule by (i) increasing the outer membrane permeability and (ii) altering the PMF (proton motive force) which is the energy-driven force of efflux pumps involved in the expulsion of toxic compounds (metabolites, drugs, antibiotics, etc.). Therefore, the combination of moxifloxacin 1 with tobramycin 2 allowed a weaker development of resistance compared to each antibiotic alone (Figure 1) [21].
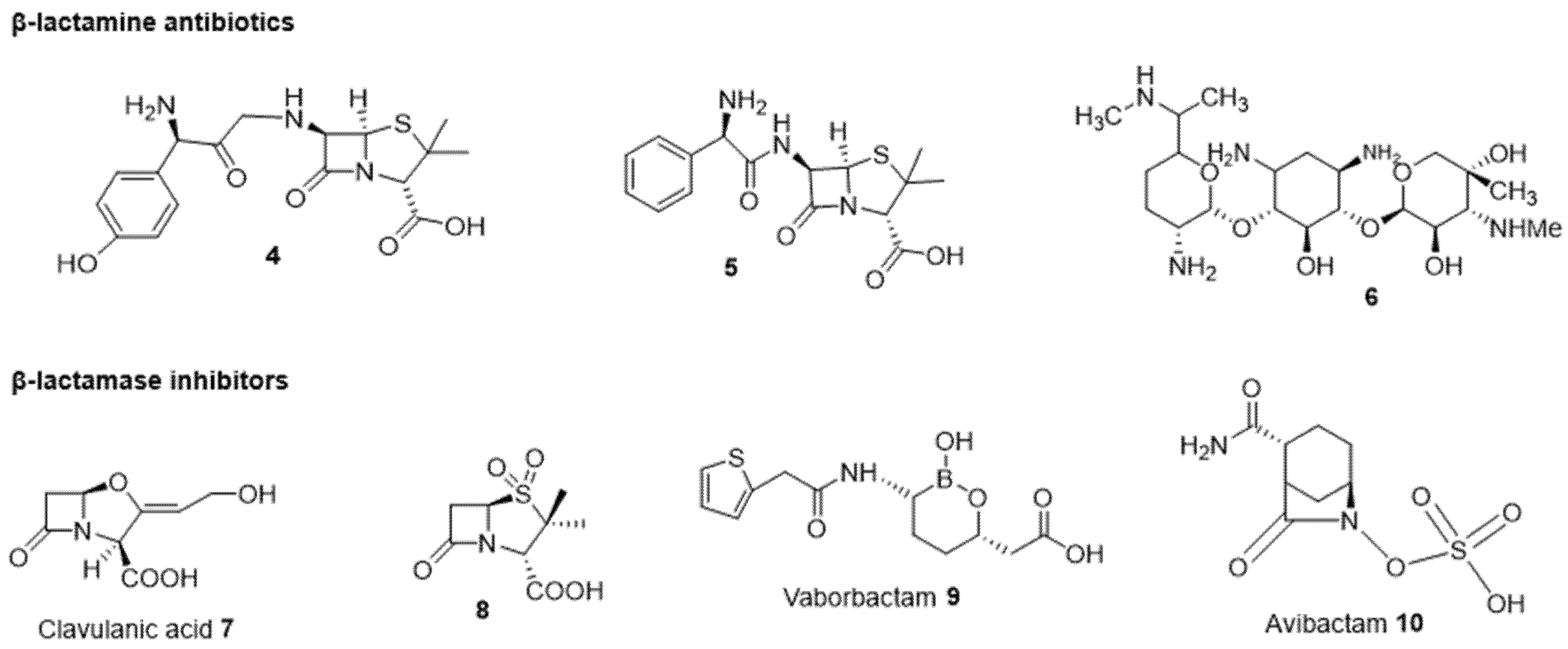
2.2.2. Antibiotic and β-Lactamase Inhibitor Combination
2.2.3. Efflux Pumps Inhibitors
Efflux pumps expel the antibiotics towards the extracellular environment and contribute to bacterial resistance. Some are selective and only expel specific substrates, while others are non-selective and expel various classes of antibiotics and non-specific substrates (detergents, organic solvents, dyes, etc.) [29]. In this case, a possible strategy consists of blocking efflux systems by so-called efflux pump inhibitors (EPIs) and therefore restoring the susceptibility of resistant strains. Several methods are used to target efflux pumps: (1) inhibit the expression of genes encoding these pumps, (2) deplete the pumps of the energy necessary for their functioning, (3) compete in the inner membrane transporter, (4) block the outlet channel in the outer membrane, (5) prevent the assembly of pump components at the membrane level [30]. For example, the use of a natural steroidal alkaloid extracted from the antidysentery plant Holarrhena, conessine 11, demonstrated a reduction in the minimum inhibitory concentrations of various antibiotics including tetracycline, cefotaxime, levofloxacin, novobiocin, rifampicin, and erythromycin and an inhibitory activity of MexAB-OprM efflux pumps (Figure 3) [31]. Epigallocatechin-3-gallate (EGCG 12) extracted from tea is another compound responsible for inhibiting this type of efflux pump in P. aeruginosa [32].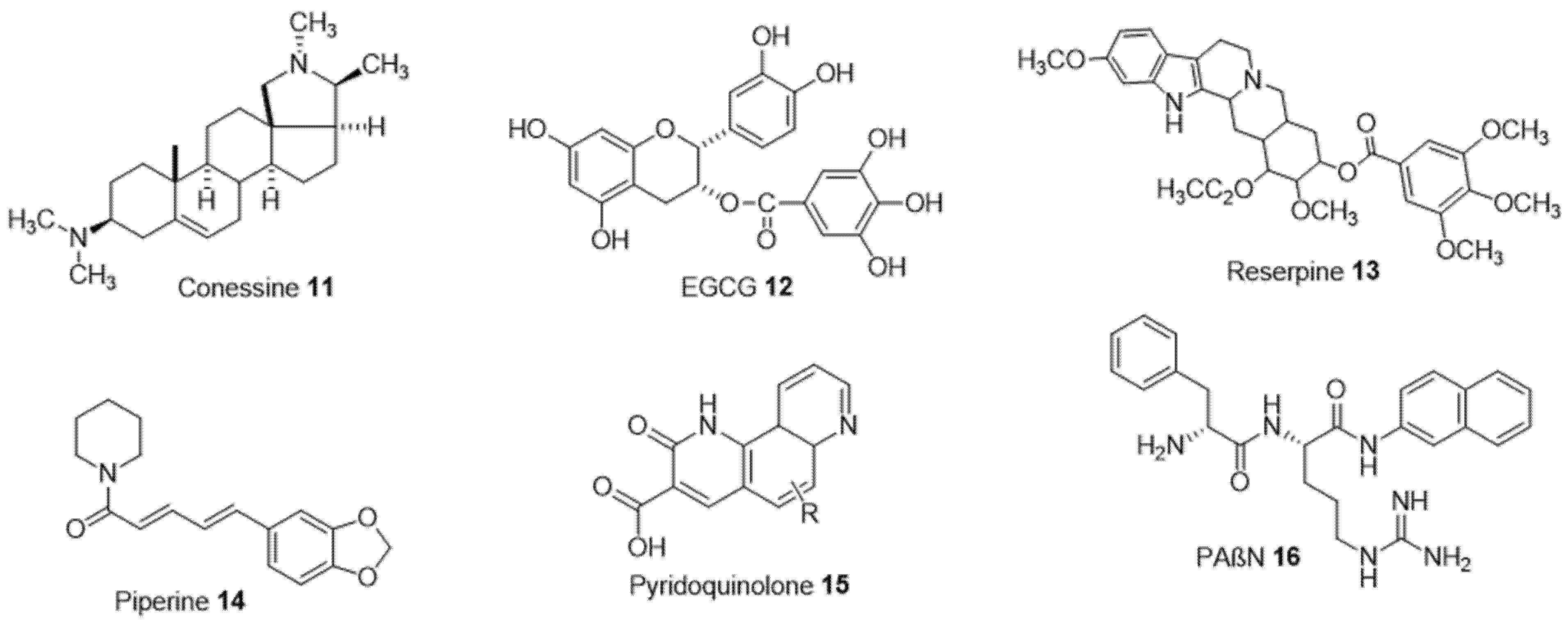
2.2.4. Liposomes Addressing Resistance
The liposomes adhere to the bacterial membrane (outer membrane) and contribute to the release of the antibiotic into the periplasmic space (Figure 4) [19]. The comparison between the Minimum Inhibitory Concentration (MIC) values of the antibiotic (tetracycline) present in a solution and in a liposome shows an important decrease (MIC in the solution is equal to 0.063 and 0.125 μg/mL for S. aureus and S. epidermidis, respectively, whereas there is a 3.9-fold decrease for S. aureus and 12.8-fold for S. epidermidis when the antibiotic is present in the liposome). This reduction indicates that the liposomes decrease the antibiotic doses required due to the fusion of the liposome structures with the bacterial membranes according to the mimetic characteristics [16][17].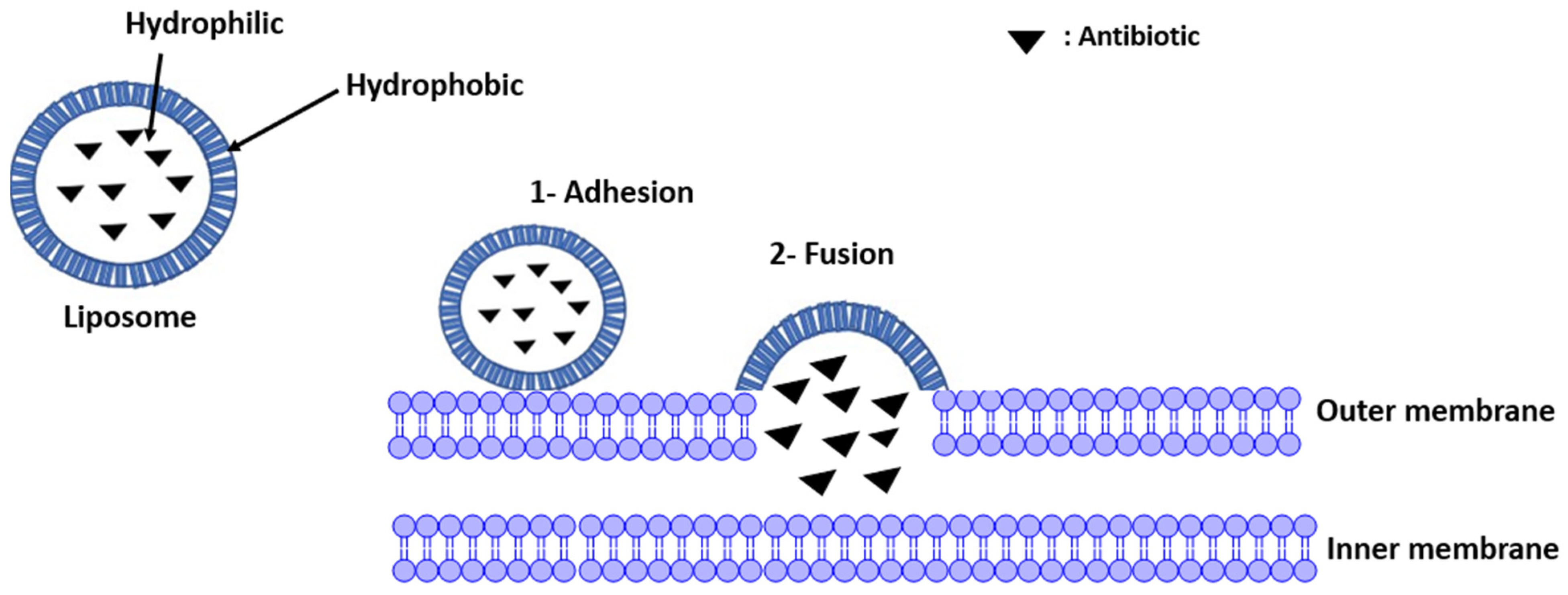
2.2.5. Potentiation of an Antibiotic Activity in the Presence of Natural Organic Compounds
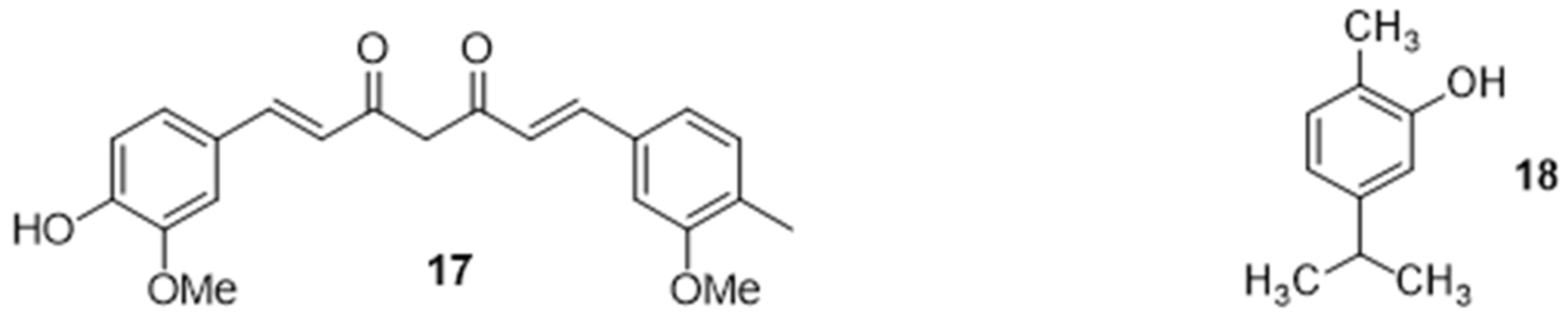
2.2.6. Potentiation of an Antibiotic Activity in the Presence of Inorganic Compounds
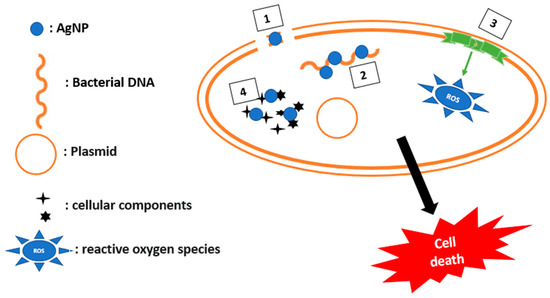
3. Outer Membrane Permeabilization
The outer membrane (OM) of Gram-negative bacteria constitutes an additional permeability barrier which protects the cell from many more antibacterial agents compared to Gram-positive bacteria. The negatively charged components of the OM interact with permeabilizers, promoting the entry of impermeable molecules. Permeabilizers are considered as potential antibiotic adjuvants to fight against bacterial resistance [29][40]. Pentamidine, an anti-protozoan drug, has shown effective activity against a wide range of Gram-negative bacteria by interacting with the lipopolysaccharide, and as a result, disrupting the outer membrane. In addition, the cationic peptide and its derivatives destroy the outer membrane by electrostatic interaction.
Menadione, a soluble synthetic vitamin converted into vitamin K2 at the intestinal level, was used to see its effect on membrane permeability against multiresistant bacteria such as S. aureus, P. aeruginosa, and E. coli. It has been demonstrated to have an antibacterial activity only against the P. aeruginosa bacterium, but when combined with antibiotics of the aminoglycoside family, it produces a synergistic effect and decreases the MICs for these antibiotics [41].
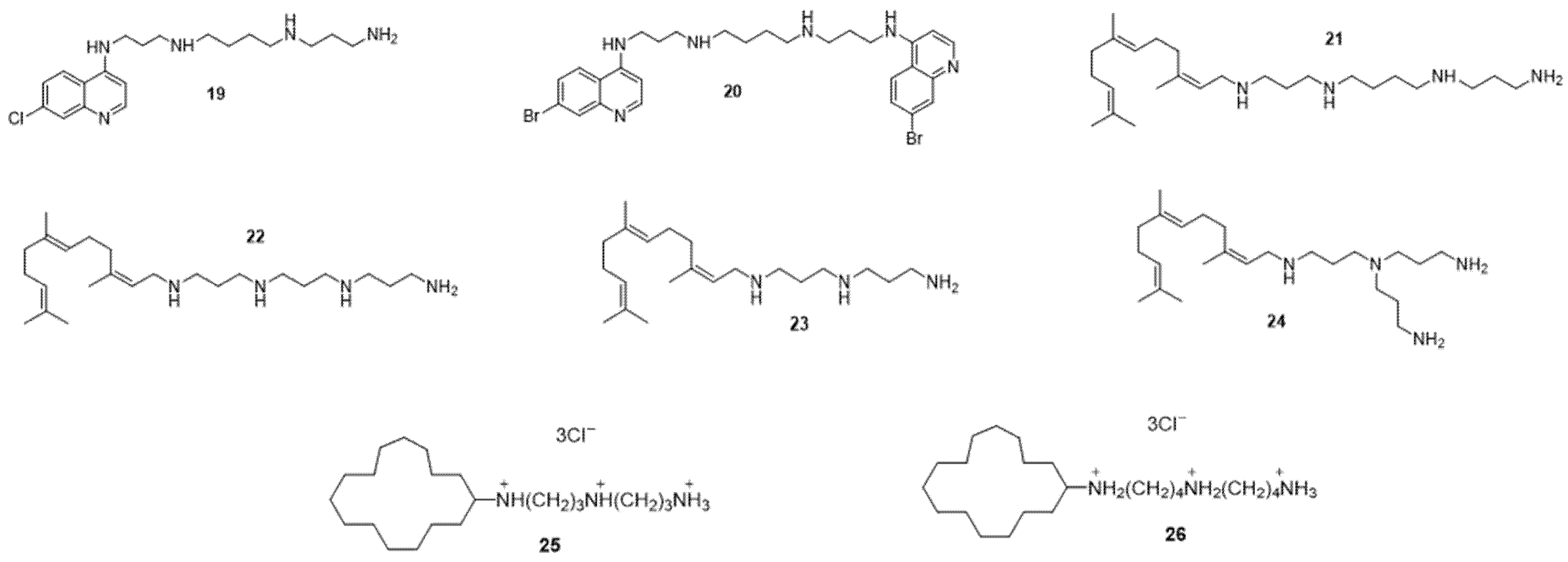
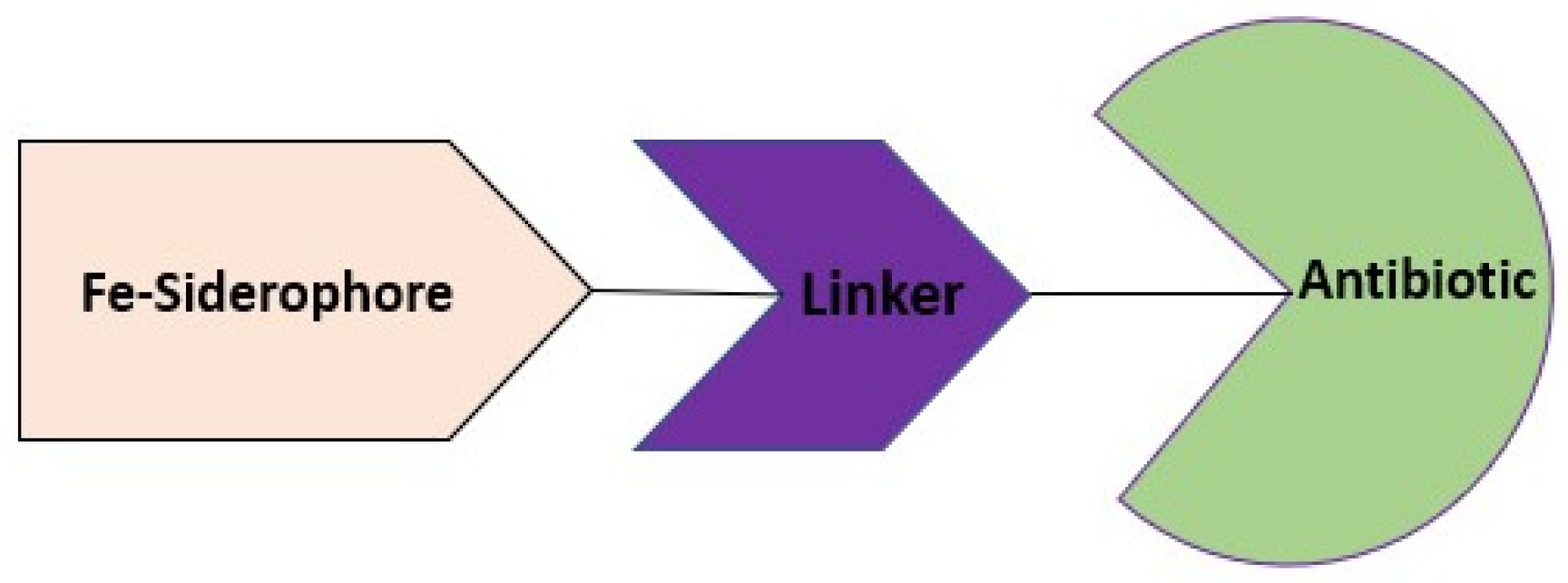
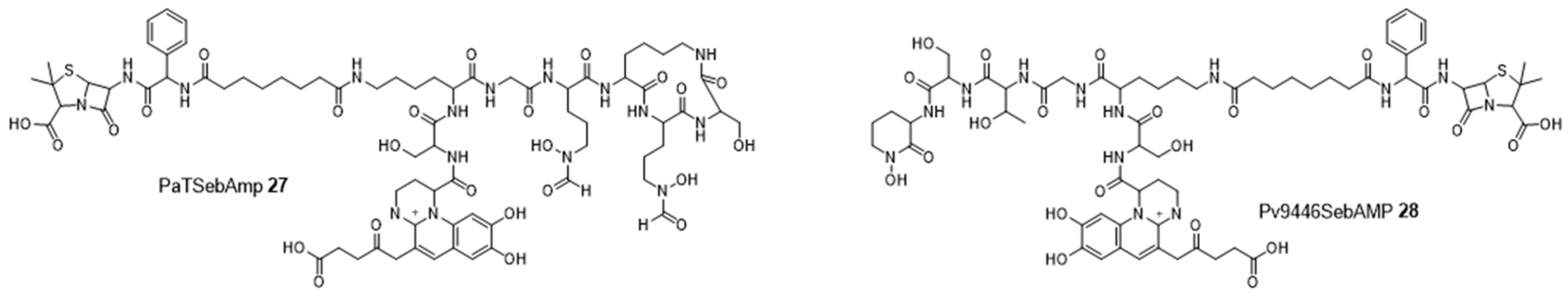
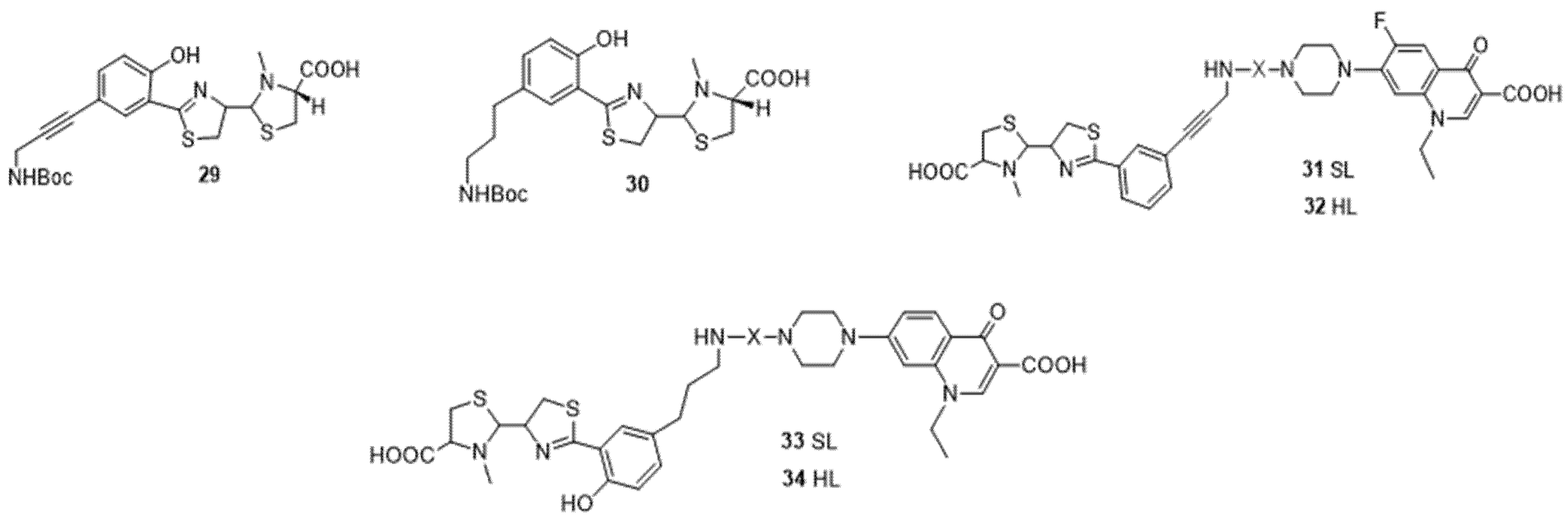
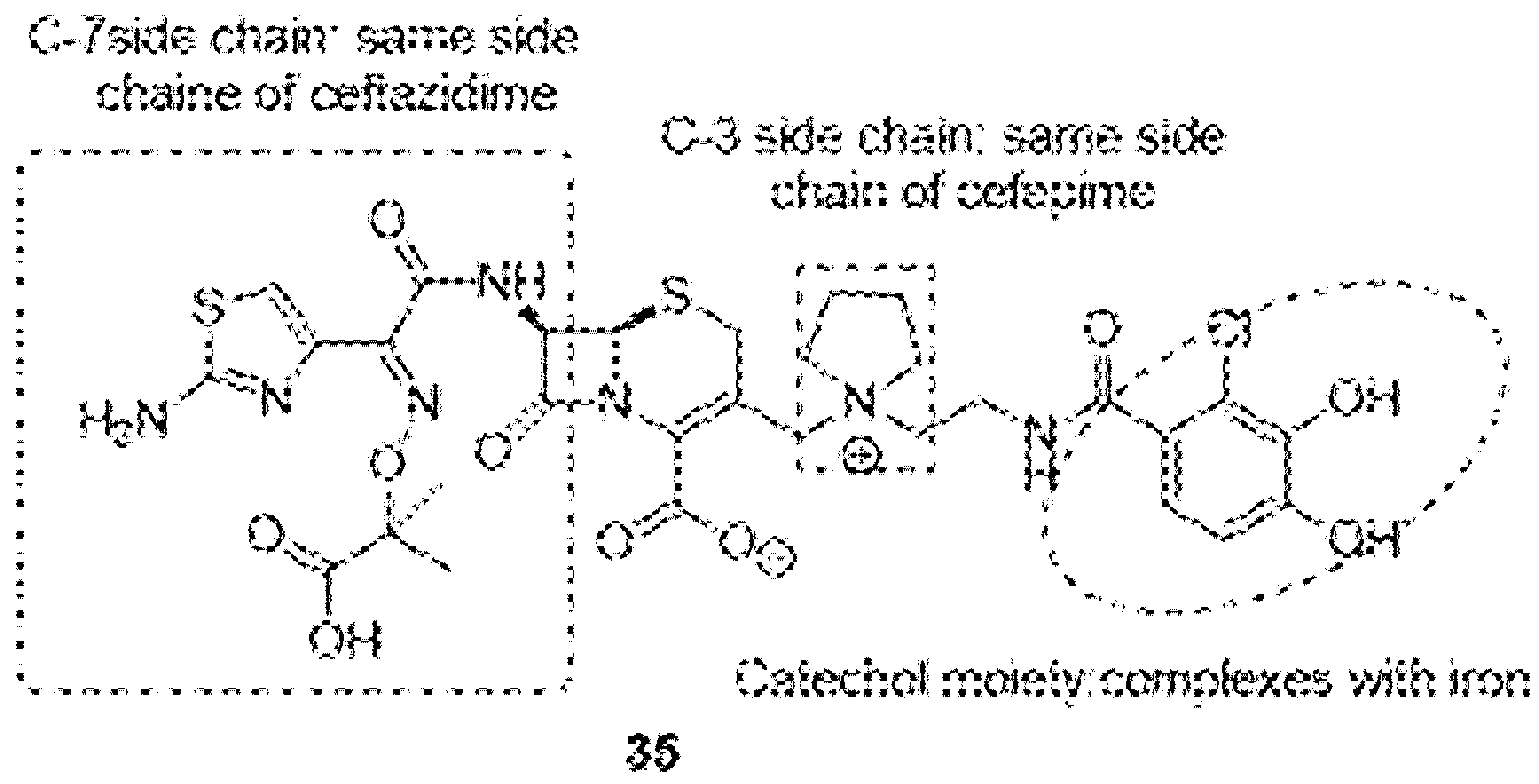
4. Cargo Delivery: Conjugation of Antibiotics with Siderophores or Sugars (The “Trojan Horse” Strategy)
4.1. Siderophores
References
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. BioMed Res. Int. 2016, 2016, 2475067.
- Antimicrobial Resistance Collaborators. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet Lond. Engl. 2022, 399, 629–655.
- Nation, R.L.; Li, J. Colistin in the 21st century. Curr. Opin. Infect. Dis. 2009, 22, 535–543.
- Coates, A.R.M.; Hu, Y.; Holt, J.; Yeh, P. Antibiotic combination therapy against resistant bacterial infections: Synergy, rejuvenation and resistance reduction. Expert Rev. Anti-Infect. Ther. 2020, 18, 5–15.
- Gadelii, A.; Hassan, K.-O.; Hakansson, A.P. Sensitizing Agents to Restore Antibiotic Resistance. In Antibiotic Drug Resistance; John Wiley & Sons, Ltd: Hoboken, NJ, USA, 2019; pp. 429–452.
- Tyers, M.; Wright, G.D. Drug combinations: A strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol. 2019, 17, 141–155.
- Murugaiyan, J.; Kumar, P.A.; Rao, G.S.; Iskandar, K.; Hawser, S.; Hays, J.P.; Mohsen, Y.; Adukkadukkam, S.; Awuah, W.A.; Jose, R.A.M.; et al. Progress in Alternative Strategies to Combat Antimicrobial Resistance: Focus on Antibiotics. Antibiotics 2022, 11, 200.
- Giordano, N.P.; Cian, M.B.; Dalebroux, Z.D. Outer Membrane Lipid Secretion and the Innate Immune Response to Gram-Negative Bacteria. Infect. Immun. 2020, 88, e00920-19.
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 464–473.
- Bajaj, H.; Scorciapino, M.A.; Moynié, L.; Page, M.G.; Naismith, J.H.; Ceccarelli, M.; Winterhalter, M. Molecular Basis of Filtering Carbapenems by Porins from β-Lactam-resistant Clinical Strains of Escherichia coli. J. Biol. Chem. 2016, 291, 2837–2847.
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300–305.
- Mutations in the Quinolone Resistance-Determining Regions of gyrA and parC in Enterobacteriaceae Isolates from Brazil—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/24031957/ (accessed on 27 November 2022).
- Lima, L.M.; da Silva, B.N.M.; Barbosa, G.; Barreiro, E.J. β-lactam antibiotics: An overview from a medicinal chemistry perspective. Eur. J. Med. Chem. 2020, 208, 112829.
- Mohr, K.I. History of Antibiotics Research. Curr. Top. Microbiol. Immunol. 2016, 398, 237–272.
- Cochrane, S.A.; Lohans, C.T. Breaking down the cell wall: Strategies for antibiotic discovery targeting bacterial transpeptidases. Eur. J. Med. Chem. 2020, 194, 112262.
- Atzori, A.; Malviya, V.N.; Malloci, G.; Dreier, J.; Pos, K.M.; Vargiu, A.V.; Ruggerone, P. Identification and characterization of carbapenem binding sites within the RND-transporter AcrB. Biochim. Biophys. Acta (BBA)—Biomembr. 2019, 1861, 62–74.
- Dashtbani-Roozbehani, A.; Brown, M.H. Efflux Pump Mediated Antimicrobial Resistance by Staphylococci in Health-Related Environments: Challenges and the Quest for Inhibition. Antibiotics 2021, 10, 1502.
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules 2020, 25, 1340.
- Pajares-Chamorro, N.; Hammer, N.D.; Chatzistavrou, X. Materials for restoring lost Activity: Old drugs for new bugs. Adv. Drug Deliv. Rev. 2022, 186, 114302.
- Ferrand, A.; Vergalli, J.; Pagès, J.-M.; Davin-Regli, A. An Intertwined Network of Regulation Controls Membrane Permeability Including Drug Influx and Efflux in Enterobacteriaceae. Microorganisms 2020, 8, 833.
- Gorityala, B.K.; Guchhait, G.; Goswami, S.; Fernando, D.M.; Kumar, A.; Zhanel, G.G.; Schweizer, F. Hybrid Antibiotic Overcomes Resistance in P. aeruginosa by Enhancing Outer Membrane Penetration and Reducing Efflux. J. Med. Chem. 2016, 59, 8441–8455.
- Huttner, A.; Bielicki, J.; Clements, M.; Frimodt-Møller, N.; Muller, A.; Paccaud, J.-P.; Mouton, J. Oral amoxicillin and amoxicillin–clavulanic acid: Properties, indications and usage. Clin. Microbiol. Infect. 2020, 26, 871–879.
- Sakulchit, T.; Goldman, R.D. Antibiotic therapy for children with acute otitis media. Can. Fam. Physician Med. Fam. Can. 2017, 63, 685–687.
- Vazouras, K.; Velali, K.; Tassiou, I.; Anastasiou-Katsiardani, A.; Athanasopoulou, K.; Barbouni, A.; Jackson, C.; Folgori, L.; Zaoutis, T.; Basmaci, R.; et al. Antibiotic treatment and antimicrobial resistance in children with urinary tract infections. J. Glob. Antimicrob. Resist. 2020, 20, 4–10.
- Espinosa-Gongora, C.; Jessen, L.R.; Kieler, I.N.; Damborg, P.; Bjørnvad, C.R.; Gudeta, D.D.; dos Santos, T.P.; Sablier-Gallis, F.; Sayah-Jeanne, S.; Corbel, T.; et al. Impact of oral amoxicillin and amoxicillin/clavulanic acid treatment on bacterial diversity and β-lactam resistance in the canine faecal microbiota. J. Antimicrob. Chemother. 2020, 75, 351–361.
- Shapiro, A.B.; Moussa, S.H.; McLeod, S.M.; Durand-Réville, T.; Miller, A.A. Durlobactam, a New Diazabicyclooctane β-Lactamase Inhibitor for the Treatment of Acinetobacter Infections in Combination with Sulbactam. Front. Microbiol. 2021, 12, 709974.
- Ogawara, H. Penicillin-binding proteins in Actinobacteria. J. Antibiot. 2015, 68, 223–245.
- Oli, A.N.; Eze, D.E.; Gugu, T.H.; Ezeobi, I.; Maduagwu, U.N.; Ihekwereme, C.P. Multi-antibiotic resistant extended-spectrum beta-lactamase producing bacteria pose a challenge to the effective treatment of wound and skin infections. Pan Afr. Med. J. 2017, 27, 66.
- Masi, M.; Réfregiers, M.; Pos, K.M.; Pagès, J.-M. Mechanisms of envelope permeability and antibiotic influx and efflux in Gram-negative bacteria. Nat. Microbiol. 2017, 2, 17001.
- Douafer, H.; Andrieu, V.; Phanstiel, O.; Brunel, J.M. Antibiotic Adjuvants: Make Antibiotics Great Again! J. Med. Chem. 2019, 62, 8665–8681.
- Siriyong, T.; Srimanote, P.; Chusri, S.; Yingyongnarongkul, B.-E.; Suaisom, C.; Tipmanee, V.; Voravuthikunchai, S.P. Conessine as a novel inhibitor of multidrug efflux pump systems in Pseudomonas aeruginosa. BMC Complement. Altern. Med. 2017, 17, 405.
- Kanagaratnam, R.; Sheikh, R.; Alharbi, F.; Kwon, D.H. An efflux pump (MexAB-OprM) of Pseudomonas aeruginosa is associated with antibacterial activity of Epigallocatechin-3-gallate (EGCG). Phytomedicine Int. J. Phytother. Phytopharm. 2017, 36, 194–200.
- Pathania, R.; Sharma, A.; Gupta, V.K. Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian J. Med. Res. 2019, 149, 129–145.
- Chevalier, J.; Atifi, S.; Eyraud, A.; Mahamoud, A.; Barbe, J.; Pagès, J.-M. New Pyridoquinoline Derivatives as Potential Inhibitors of the Fluoroquinolone Efflux Pump in Resistant Enterobacter aerogenes Strains. J. Med. Chem. 2001, 44, 4023–4026.
- Pradel, E.; Pagès, J.-M. The AcrAB-TolC Efflux Pump Contributes to Multidrug Resistance in the Nosocomial Pathogen Enterobacter aerogenes. Antimicrob. Agents Chemother. 2002, 46, 2640–2643.
- Marini, E.; Di Giulio, M.; Magi, G.; Di Lodovico, S.; Cimarelli, M.E.; Brenciani, A.; Nostro, A.; Cellini, L.; Facinelli, B. Curcumin, an antibiotic resistance breaker against a multiresistant clinical isolate of Mycobacterium abscessus. Phytother. Res. 2018, 32, 488–495.
- Balderrama-González, A.-S.; Piñón-Castillo, H.-A.; Ramírez-Valdespino, C.-A.; Landeros-Martínez, L.-L.; Orrantia-Borunda, E.; Esparza-Ponce, H.-E. Antimicrobial Resistance and Inorganic Nanoparticles. Int. J. Mol. Sci. 2021, 22, 12890.
- Bruna, T.; Maldonado-Bravo, F.; Jara, P.; Caro, N. Silver Nanoparticles and Their Antibacterial Applications. Int. J. Mol. Sci. 2021, 22, 7202.
- Schuhladen, K.; Mukoo, P.; Liverani, L.; Neščáková, Z.; Boccaccini, A.R. Manuka honey and bioactive glass impart methylcellulose foams with antibacterial effects for wound-healing applications. Biomed. Mater. 2020, 15, 065002.
- Vergalli, J.; Bodrenko, I.V.; Masi, M.; Moynié, L.; Acosta-Gutierrez, S.; Naismith, J.H.; Davin-Regli, A.; Ceccarelli, M.; Berg, B.V.D.; Winterhalter, M.; et al. Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria. Nat. Rev. Genet. 2020, 18, 164–176.
- Andrade, J.C.; Braga, M.F.B.M.; Guedes, G.M.M.; Tintino, S.R.; Freitas, M.A.; Quintans, L.J.; Menezes, I.R.; Coutinho, H.D. Menadione (vitamin K) enhances the antibiotic activity of drugs by cell membrane permeabilization mechanism. Saudi J. Biol. Sci. 2017, 24, 59–64.
- Troïa, T.; Siad, J.; Di Giorgio, C.; Brunel, J.M. Design and synthesis of new polyamine quinoline antibiotic enhancers to fight resistant gram-negative P. aeruginosa bacteria. Eur. J. Med. Chem. Rep. 2022, 5, 100054.
- Troudi, A.; Douafer, H.; Bolla, J.-M.; Klibi, N.; Brunel, J.M. Antibiotic Adjuvants to Rescue Pseudomonas aeruginosa from Tetracycline Antibiotics Resistance. Anti-Infect. Agents 2021, 19, 110–116.
- Wang, G.W.; Brunel, J.-M.; Bolla, J.-M.; Van Bambeke, F. The Polyaminoisoprenyl Potentiator NV716 Revives Old Disused Antibiotics against Intracellular Forms of Infection by Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2021, 65, e02028-20.
- Borselli, D.; Blanchet, M.; Bolla, J.-M.; Muth, A.; Skruber, K.; Iv, O.P.; Brunel, J.M. Motuporamine Derivatives as Antimicrobial Agents and Antibiotic Enhancers against Resistant Gram-Negative Bacteria. Chembiochem Eur. J. Chem. Biol. 2017, 18, 276–283.
- Soriano, A.; Carmeli, Y.; Omrani, A.S.; Moore, L.S.P.; Tawadrous, M.; Irani, P. Ceftazidime-Avibactam for the Treatment of Serious Gram-Negative Infections with Limited Treatment Options: A Systematic Literature Review. Infect. Dis. Ther. 2021, 10, 1989–2034.
- Kramer, J.; Özkaya, Ö.; Kümmerli, R. Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 2020, 18, 152–163.
- Swayambhu, G.; Bruno, M.; Gulick, A.M.; Pfeifer, B.A. Siderophore natural products as pharmaceutical agents. Curr. Opin. Biotechnol. 2021, 69, 242–251.
- Two Novel Proteins, TtpB2 and TtpD2, are Essential for Iron Transport in the TonB2 System of Vibrio Vulnificus—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/31595707/ (accessed on 25 June 2022).
- Page, M.G.P. The Role of Iron and Siderophores in Infection, and the Development of Siderophore Antibiotics. Clin. Infect. Dis. 2019, 69, S529–S537.
- Möllmann, U.; Heinisch, L.; Bauernfeind, A.; Köhler, T.; Ankel-Fuchs, D. Siderophores as drug delivery agents: Application of the “Trojan Horse” strategy. Biometals Int. J. Role Met. Ions Biol. Biochem. Med. 2009, 22, 615–624.
- Wang, M.; Zhang, Y.; Lv, L.; Kong, D.; Niu, G. Biosynthesis and Chemical Synthesis of Albomycin Nucleoside Antibiotics. Antibiotics 2022, 11, 438.
- Hartmann, A.; Fiedler, H.P.; Braun, V. Uptake and Conversion of the Antibiotic Albomycin by Escherichia Coli K-12. Eur. J. Biochem. 1979, 99, 517–524.
- Mislin, G.L.A.; Schalk, I.J. Siderophore-dependent iron uptake systems as gates for antibiotic Trojan horse strategies against Pseudomonas aeruginosa. Met. Integr. Biometal Sci. 2014, 6, 408–420.
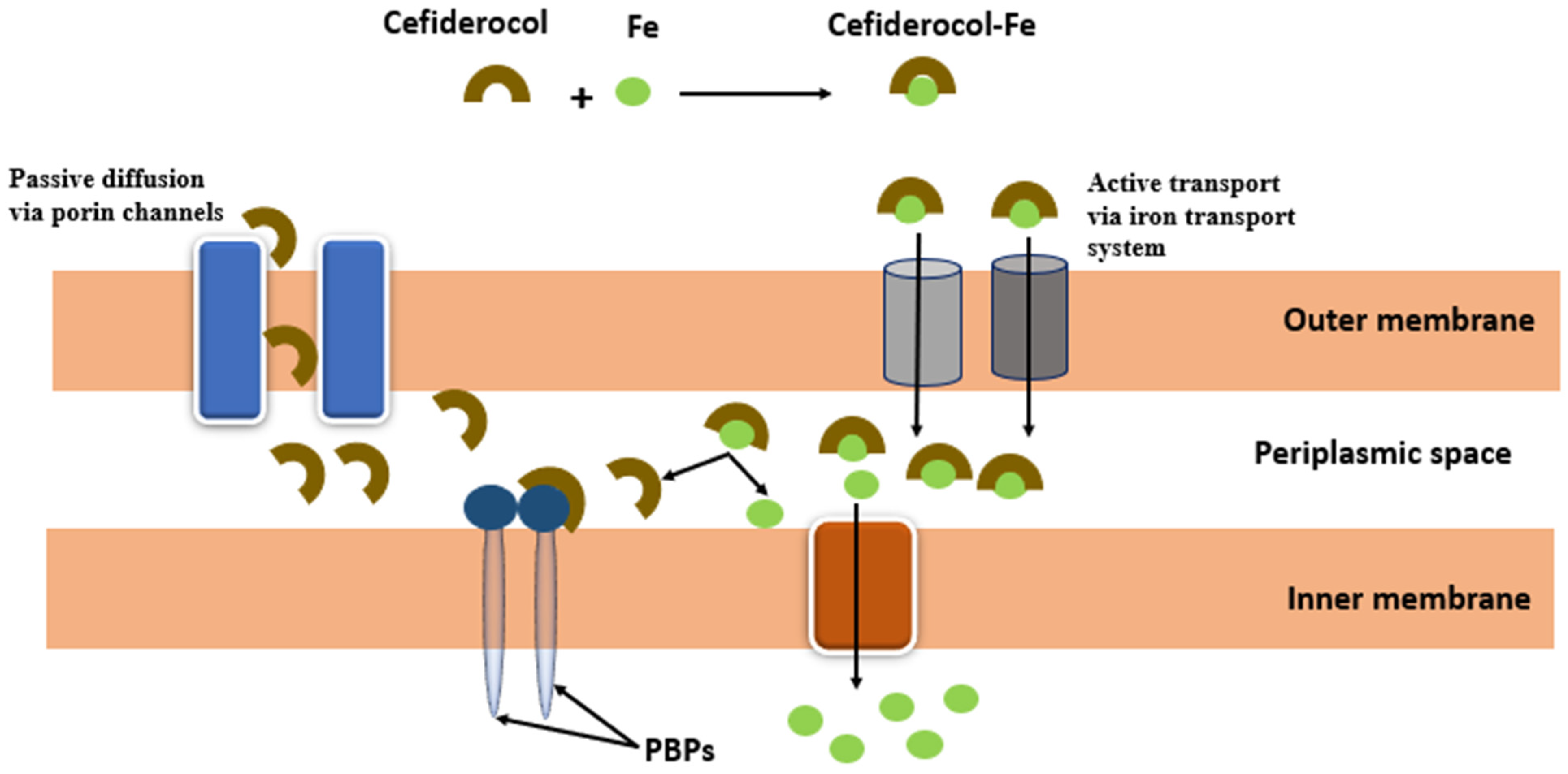
- El-Lababidi, R.M.; Rizk, J.G. Cefiderocol: A Siderophore Cephalosporin. Ann. Pharmacother. 2020, 54, 1215–1231.
- Zhanel, G.G.; Golden, A.R.; Zelenitsky, S.; Wiebe, K.; Lawrence, C.K.; Adam, H.J.; Idowu, T.; Domalaon, R.; Schweizer, F.; Zhanel, M.A.; et al. Cefiderocol: A Siderophore Cephalosporin with Activity Against Carbapenem-Resistant and Multidrug-Resistant Gram-Negative Bacilli. Drugs 2019, 79, 271–289.
