Recent studies have identified numerous m6A-regulated genes, including glucose and lipid metabolism-related enzymes, transcription factors, and signaling pathways, which exerted important effects on hyperglycemia, insulin signaling transduction, lipid accumulation, and plaque formation.
2. m6A Methylation
M6A mainly refers to the methylation of the sixth nitrogen of adenosine, which accounts for the most abundant internal modification that occurs in eukaryotic mRNA. Other than mRNA, m6A occurs in long non-coding RNA (lncRNA), circular RNA (circRNA), and microRNA (miRNA)
[13][14][15][13,14,15]. The highly conserved and dynamic m6A modification level has been established to be regulated by methyltransferases (termed “writers”) and demethylases (termed “erasers”) (
Table 1). The m6A writers comprised Methyltransferase-like protein 3 (METTL3), methyltransferase-like protein 14 (METTL14)
[16], methyltransferase-like protein 16 (METTL16), Wilms tumor 1-associated protein (WTAP), Vir-like m6A methyltransferase associated (VIRMA/KIAA1429), RNA-binding motifs protein15/15B (RBM15/15B), and Zinc Finger CCCH-Type Containing 13 (ZC3H13). Typically, METTL3, METTL14, and WTAP form a methyltransferase complex (MTC), which recognizes a consensus RNA sequence RRACH (R = G or A; H = A, C or U) and catalyzes the m6A modification on mRNAs, while METTL16 regulates S-adenosylmethionine (SAM) homeostasis
[17]. VIRMA recruits MTC to specific mRNA regions and interacts with cleavage and polyadenylation specific factor 5/6 (CPSF5/6)
[18]. RBM15/15B mainly guides METTL3-METTL14 heterodimer to uracil U-rich RNA sites for methylation
[19]. ZC3H13 bridges WTAP to the mRNA binding factor Nito and contributes to the nuclear localization of MTC
[20]. Additionally, IME4 and MUM2 mediate m6A modification of yeast mRNA
[21].
Table 1.
The biological function of m6A regulators in RNA metabolism.
The discovery of m6A demethylases, including fat mass and obesity-associated (FTO) and ALKBH5, verify m6A methylation as a dynamic and reversible RNA modification and thus are regarded as m6A erasers. Both FTO and ALKBH5 belong to the alpha-ketoglutarate-dependent dioxygenase family. FTO promotes mRNA splicing and translation
[22], while ALKBH5 mainly promotes mRNA nuclear export, mRNA splicing, and long 3′-UTR mRNA production by clearing m6A
[23].
The m6A binding proteins, referred to as “Readers”, specifically recognize and bind with the m6A-modified mRNA to regulate gene expression via impacting mRNA transcription, stability, splicing, or nuclear export. The most important m6A readers are YTH domain-containing family proteins, including YTHDF1/2/3 and YTHDC1/2. YTHDF1 promotes mRNA translation and protein synthesis, and YTHDF2 reduces mRNA stability and regulates mRNA localization; YTHDF3 interacts with YTHDF1 to promote mRNA translation or assists YTHDF2-mediated RNA degradation
[24]. Nuclear YTHDC1 mediates RNA splicing, export, and transcriptional silencing
[25]. YTHDC2 mainly promotes the translation of target RNA but reduces their abundance
[26]. In addition, eukaryotic translation initiation factor 3 (eIF3) promotes mRNA translation by recruiting ribosomal initiation complexes
[27]. Insulin-like growth factor 2 mRNA binding proteins (IGF2BPs) enhance the stability of target transcripts, and the heterogeneous nuclear ribonucleoprotein (HNRNP) family mainly mediates mRNA splicing
[28][29][28,29]. Among these, HNRNPA2B1 regulates alternative splicing and primary microRNA processing, and HNRNPC/G mediates pre-mRNA splicing and processing.
3. m6A Modification and Glucose Metabolism
Glucose metabolism involves a very complex regulatory network, including anaerobic glycolysis, aerobic oxidation, pentose phosphate pathway, glycogen synthesis, and gluconeogenesis
[30]. An increasing number of studies have reported that m6A modification is an important regulatory mechanism of glucose homeostasis and downstream effects (
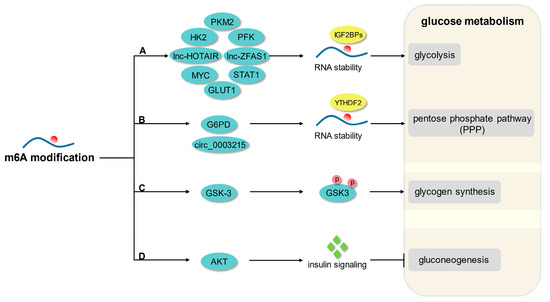
Figure 1).
Figure 1. The principal mechanism of m6A modification in glucose metabolism. (A) M6A modification promotes glycolysis by enhancing the mRNA stability of GLUT1, HK2, PFK, STAT1, PKM2, MYC, lnc-HOTAIR, and lnc-ZFAS1. (B) M6A modification enhances PPP flux by mediating the mRNA stability of G6PD and circ_0003215. (C) M6A modification regulates glycogen synthesis by mediating phosphorylation of GSK-3. (D) M6A modification modulates gluconeogenesis in response to AKT-mediated insulin signaling.
3.1. Glycolysis
Glycolysis is a pivotal energy-producing pathway in organisms, which decomposes glucose to pyruvate under anaerobic conditions, with the release of free energy into adenosine triphosphate (ATP). Typically, monosaccharides are transported to the cytoplasm by glucose transporters (GLUTs) or sodium-dependent glucose cotransporters (SGLTs), then undergo the preparation phase of glucose activation and cleavage and the release energy phase of oxidative phosphorylation, which are closely related to hexokinase (HK), phosphofructokinase-1 (PFK-1) and pyruvate kinase (PK), and ultimately generate pyruvate and ATP
[31].
A growing number of studies have uncovered the broad effects of m6A modification on metabolic networks by regulating glycolytic genes and signaling pathways. METTL3/IGF2BP2-mediated m6A modification has been reported to promote glycolysis by enhancing the stability of HK2 and GLUT1
[32][33][32,33]. METTL14-mediated m6A modification not only promotes glycolysis by attenuating sirtuin6 (SIRT6) stability but also facilitates hypoxia-inducible factor 1 subunit alpha (HIF1A)-mediated glycolysis and cell proliferation by inhibiting the expression of phosphatase LHPP
[34][35][34,35]. WTAP enhances glycolytic activity by mediating m6A methylation of HK2 and enolase 1 (ENO1) mRNA
[36][37][38][36,37,38]. The m6A methyltransferase KIAA1429 positively regulates aerobic glycolysis in a GLUTI or HK2-dependent manner
[39][40][39,40], and ZC3H13 facilitates glycolysis by enhancing the stability of pyruvate kinase M2 (PKM2) mRNA
[41]. In addition, FTO-mediated demethylation promotes HK2 and PKM2-mediated glycolysis via upregulating the expression of lncRNA HOTAIR and autophagy associated 5 gene (ATG5), respectively
[42][43][42,43], while down-regulation of FTO participates MYC-mediated cellular glycolysis and the regulation of IL-6/JAK2/STAT3 signaling pathways
[44][45][44,45]. FTO/YTHDF2 mediates post-transcriptional upregulation of phosphofructokinase platelet (PFKP) and lactate dehydrogenase B (LDHB) and activates aerobic glycolysis
[46]. ALKBH5 is involved in the regulation of casein kinase 2 (CK2) α-mediated glycolysis in an m6A-dependent manner
[47]. M6A reader IMP2 enhances the stability of lncRNA ZFAS1 and facilitates the exposure of ATP-binding sites, thereby accelerating ATP hydrolysis and glycolysis
[48]. IGF2BP1/2 is involved in the regulation of MYC-mediated glycolysis and provides an additional energy source for cell metabolism
[49][50][51][49,50,51]. In cellular metabolism, pyruvate dehydrogenase kinase 4 (PDK4) methylation can be recognized by YTHDF1/eEF-2 complex and IGF2BP3 to direct carbon flux from oxidative phosphorylation (OXPHOS) to glycolysis
[52]. The interaction of YTHDF2 with RNA-binding motif protein 4 (RBM4) promotes signal transduction and activator of transcription 1 (STAT1)-mediated glycolysis, which participates in regulating macrophage polarization and inflammatory factor expression
[53].
3.2. Pentose Phosphate Pathway
The pentose phosphate pathway (PPP), also known as the hexose phosphate bypass, is generally divided into two branches: oxidative and non-oxidative. During the highly active oxidation phase in most eukaryotes, glucose-6-phosphate (G-6-P) is converted to ribulose-5-phosphate, carbon dioxide, and nicotinamide adenine dinucleotide phosphate (NADPH)
[54]. The non-oxidative branch is nearly ubiquitous and supports the nucleic acid skeleton and aromatic amino acid biosynthesis by increasing the expression of 5-phosphate ribose and erythritol-4-phosphate
[55][56][55,56].
As a key enzyme of PPP, glucose-6-phosphate dehydrogenase (G6PD) is overactive in metabolic pathways and participates in the regulation of redox status. Recent studies have demonstrated that tumor cells respond to chemotherapy-induced reactive oxygen species (ROS) accumulation by activating PPP to increase NADPH, thereby adapting to oxidative stress and maintaining malignant cell proliferation
[57]. In a glioma, ALKBH5 enhances the stability of G6PD mRNA and PPP flux by eliminating m6A methylation
[58]. YTHDF2 facilitates mRNA translation of G6PD in an m6A-dependent manner, thereby enhancing PPP activity and tumor cell viability
[59]. In addition, YTHDF2 promotes the miR-663b/DLG4/G6PD axis and pentose phosphate pathway by mediating circ_0003215 RNA degradation
[60].
3.3. Glycogen Synthesis and Gluconeogenesis
As a storage form of sugar, glycogen synthesis refers to the process of converting activated glucose into glycogen under the catalysis of glycogen synthase. Glycogen synthase activity is regulated by phosphorylation/de-phosphorylation of various serine/threonine kinases, among which glycogen synthase kinase 3 (GSK-3) is widely involved in the physiological and metabolic processes. Under normal feeding conditions, enhanced insulin signaling activates protein kinase B (AKT), which then inactivates GSK-3 through phosphorylation and ultimately promotes glycogen synthesis in response to increased glucose uptake. In contrast, fasting activates GSK-3 by de-phosphorylation, which inhibits glycogen synthesis and facilitates glycogenolysis, supplying the body with fuel reserve
[61]. As a highly conserved negative regulator of receptor tyrosine kinases, cytokines, and Wnt signaling pathways, GSK-3 participates in the m6A methylation regulatory network and affects multiple downstream effectors. Similarly, downregulated microRNA-6125 and hypoxia-induced lncRNA STEAP3-AS1 interact competitively with YTHDF2, which results in phosphorylation and inactivation of GSK3β, activation of Wnt/β-catenin signaling pathways, and glycogen synthesis
[62][63][62,63]. METTL14 negatively regulates the expression of fibroblast growth factor receptor 4 (FGFR4) in an m6A-dependent manner, while FGFR4 activates glycogen synthesis and the β-catenin/TCF-4 signaling pathway through phosphorylation of GSK-3β
[64]. M6A mediates phosphorylation of AKT/GSK-3β and activation of tensin homolog protein (PTEN), which promotes glycogen synthesis and protects neurons from pyroptosis induced by cerebral ischemia/reperfusion (I/R)
[65]. Cardiac hypertrophy-associated PIWI-interacting RNA (CHAPIR) competitively binds METTL3 and blocks m6A methylation of polymerase family member 10 (PARP10), while up-regulation of PARP10 facilitates glycogen synthesis and pathological cardiac hypertrophy by inhibiting the kinase activity of GSK-3β
[66].
The process by which organisms synthesize glucose or glycogen from non-sugar precursors such as lactic acid, glycerol, and glycogenic amino acids is known as gluconeogenesis. Under starvation, the liver promotes gluconeogenesis by decreasing insulin concentration and increasing glucagon concentration, which is the main cause of diabetic hyperglycemic phenotype
[67]. METTL14 deficiency leads to decreased β-cell mass and insulin secretion, but β-cell-specific knockdown of METTL14 enhances insulin signaling and reduces hepatic gluconeogenesis under a high-fat diet (HFD), thus improving insulin sensitivity with compensated
[68]. This suggests that m6A methylation regulates the gluconeogenic flux in response to insulin signaling, which is of great significance for glucose homeostasis.