Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Andrei A Deviatkin.
The mRNA platform has become the method of choice in vaccine development to find new ways to fight infectious diseases. This instability is due to the fact that the five-prime and three-prime ends of the mRNA are a substrate for the ubiquitous exoribonucleases. To address the problem, circular mRNAs have been proposed for transgene delivery as they lack these ends. Notably, circular RNAs do not have a capped five-prime end, which makes it impossible to initiate translation canonically.
- circRNA
- cap-independent translation
- mRNA vaccines
1. Introduction
The more frequent and severe the infection, the more resources are invested into research on the interaction of the pathogen with the human organism. As a result, there is a need to develop effective approaches to treat and prevent disease. The most effective and safest method to prevent the spread of an infectious disease is vaccination. At the beginning of the COVID-19 pandemic, many types of vaccines were effectively used for controlling various diseases [1]. The development of alternative approaches was hampered to some extent, as there did not appear to be a problem that needed solving.
The emergence of a new challenge became a turning point in vaccine development. Indeed, there were virtually no clinical trials of mRNA vaccines to prevent infections before the COVID-19 pandemic (Figure 1, magenta line). At the same time, the number of mRNA-based vaccine trials against COVID-19 in the eleven months of 2022 does not exceed the same indicator for 2021 (Figure 1, cyan line). In contrast, the development of such drugs to prevent other infections almost doubled in one year (Figure 1, blue line). In other words, the mRNA vaccine platform that has been promoted to combat COVID-19 is increasingly being used as an approach to prevent infections in general.
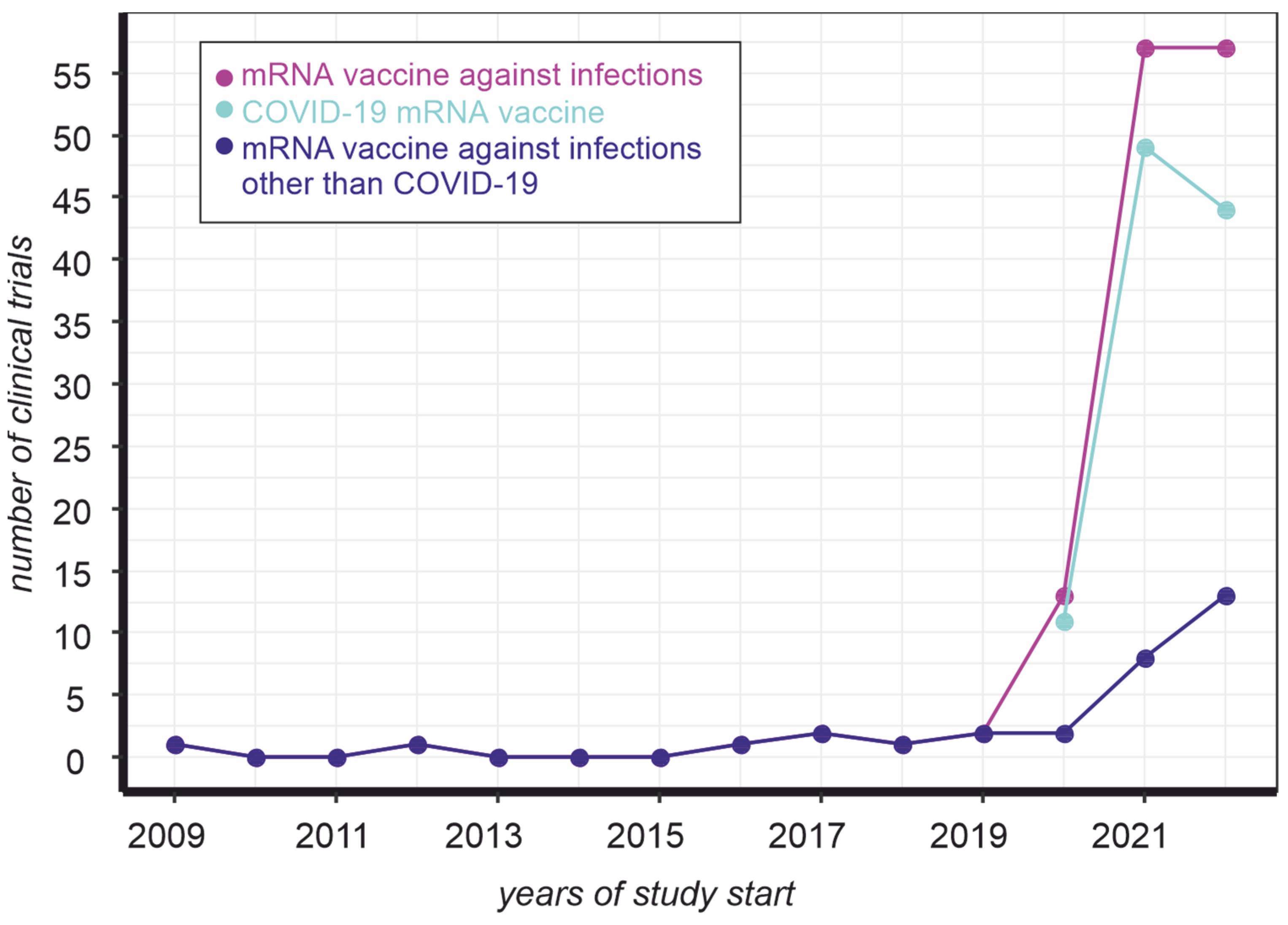
Figure 1. Dynamics of mRNA vaccine clinical trials registered on clinicaltrials.gov. The number of trials was counted by keyword ([condition “Infections”, title “mrna vaccine”] (magenta), [condition “Infections”, other terms “NOT coronavirus NOT COVID19 NOT BNT162b2”, title “mrna vaccine”] (blue), [condition “Infections”, other terms “coronavirus OR COVID19 OR BNT162b2”, title “mrna vaccine”] (cyan)) occurrence, as of 28 November 2022.
However, mRNA technology has some weaknesses. Firstly, widely used mRNA vaccines do not maintain their stability at room temperature for more than 24 h [2]. Refrigeration at +4 degrees Celsius extends the shelf life of the vaccine by several weeks, while −20 degrees Celsius extends it by several months. Therefore, there is a clear need for a second-generation mRNA-based platform for vaccine development with increased stability to ensure adequate protection against future pandemics [2].
Secondly, the half-life of mRNA in the mammalian cell is estimated to be several minutes to several days, with a median value of 7 h [3]. Surprisingly, to ourthe knowledge, the half-life of Moderna and Pfizer/BioNTech mRNA vaccines in human cells is unclear. This parameter influences the vaccine dose required to elicit a sufficient immune response.
The main reason for the instability of mRNA is its sensitivity to RNases. Exoribonucleases actively degrade the 5′ and 3′ ends of mRNA and play an important role in mRNA degradation [4]. Circular RNAs (circRNA) do not have 5′ and 3′ ends, which increases their stability compared to the linear form, which is one of its advantages. They are generally stable in serum, with a 24-h incubation at room temperature having minimal effect on titer [5]. In vivo, circRNA can remain in the cell for more than 48 h as they are resistant to degradation by exonucleases and localize mainly in the cytoplasm [6]. As such, they are a promising alternative to linear RNA for use in vaccines, as they do not require special equipment for storage. At the same time, to the best of ourthe knowledge, currently, there are no registered clinical trials of vaccines against infectious diseases based on the circular mRNA platform.
There are several ways to synthesize circRNAs. These molecules can be produced directly in cells by transcription and back-splicing when delivered as vector DNA (e.g., AAV [7]). On the other hand, circRNAs can be synthesized in vitro [8,9][8][9]. Currently, there are three methods for circRNA vaccines in in vitro production: chemical-, enzyme-, and ribozyme-based ligations. At the same time, these approaches have shortcomings (reviewed in [10]). Therefore, new protocols for large-scale production are needed to enable the cost-effective design and production of circRNA vaccines.
Much of genetic construction is tailored to canonical translation, which begins with cap-dependent initiation. Endless circular RNAs have no cap necessary for canonical translation initiation (Figure 2, left upper panel). As far as wresearchers know, only one of the possible methods of cap-independent translation initiation is used in the development of modern circular mRNA vaccines, although there are several approaches to increase the effectiveness of transgene production. The methods of translation initiation applicable to the engineering of circular mRNAs (Figure 2).
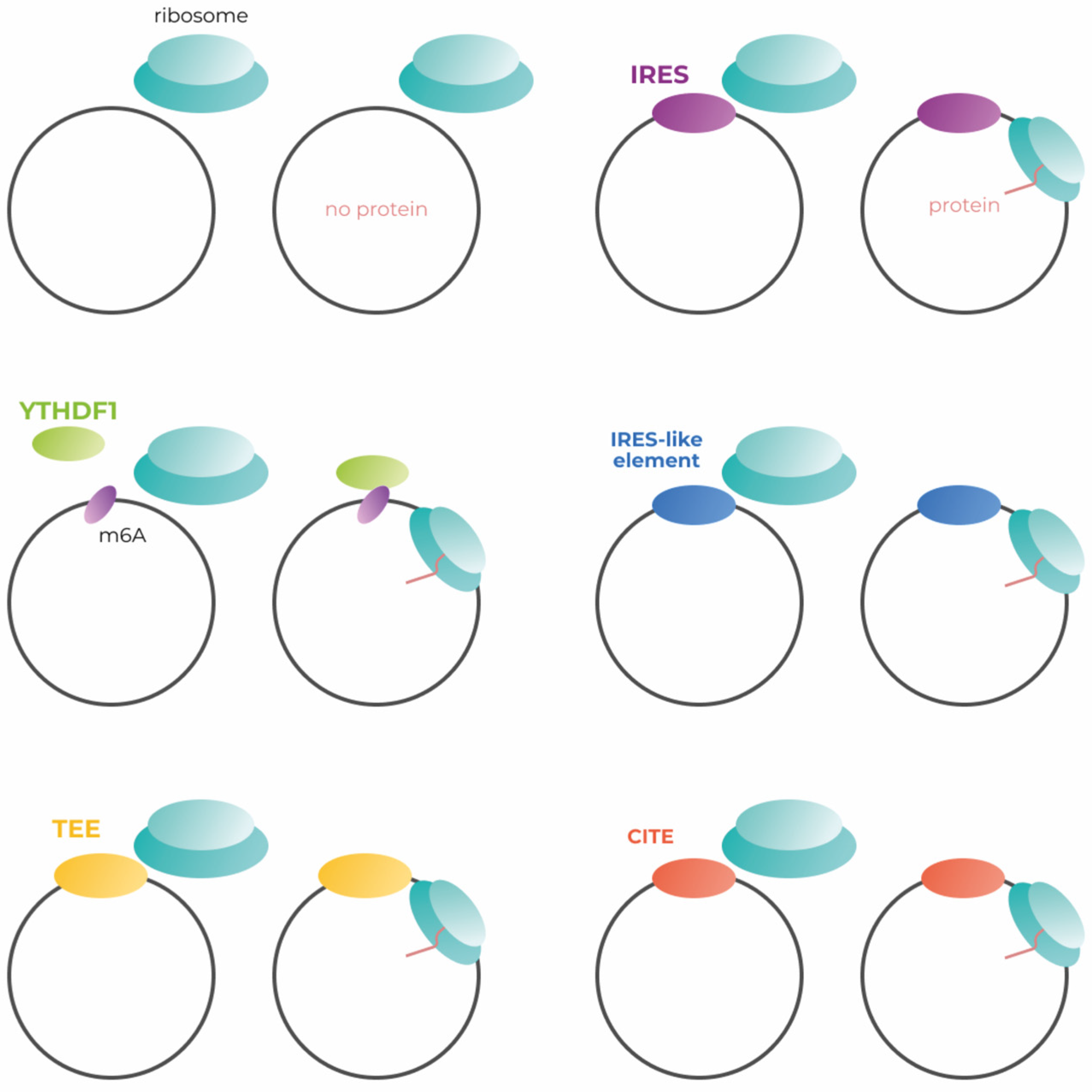
Figure 2. Simplified scheme of the mechanisms for cap-independent initiation of circRNA translation. IRES—internal ribosome entry site; m6A—N6-methyladenosine; YTHDC1—the reader of m6A modification; TEE—translation enhancing element; CITE—cap-independent translation enhancer.
2. Translation Initiation Mechanisms
For eukaryotic translation (Figure 3), the 80S ribosome should be assembled at the mRNA. The canonical cap-dependent process begins with the interaction of a small 40S ribosomal subunit with a ternary complex consisting of trimeric eukaryotic initiation factor 2 (eIF2), GTP and initiator transport RNA (Met-tRNAi) to form the 43S pre-initiation complex (PIC) [11,12][11][12]. 43S PIC binds to the 5′ end of the mRNA, interacting with the eIF4F complex (cap-binding eIF4E, the scaffolding eIF4G, and the helicase eIF4A). It should be noted that in the absence of the cap, the above complex does not bind to the 5′ end of the mRNA and translation would not be initiated in this way. 43S PIC and eIF4F complexes, together with poly(A)-binding proteins (PABPs) at the poly(A) tail, form the 48S PIC, which then scans a 5′-untranslated region in the 5′-3′ direction for the AUG start codon within the Kozak sequence context. Start codon recognition by methionine anticodon triggers GTP hydrolysis and eIF2-GDP release, as well as 60S ribosomal subunit interaction mediated by eIF5B to form the 80S initiation complex [13]. eIF1, eIF1A, eIF2, eIF3, eIF4B, eIF4F, eIF5, eIF5B, and PABP are the key factors involved in cap-dependent translation initiation.
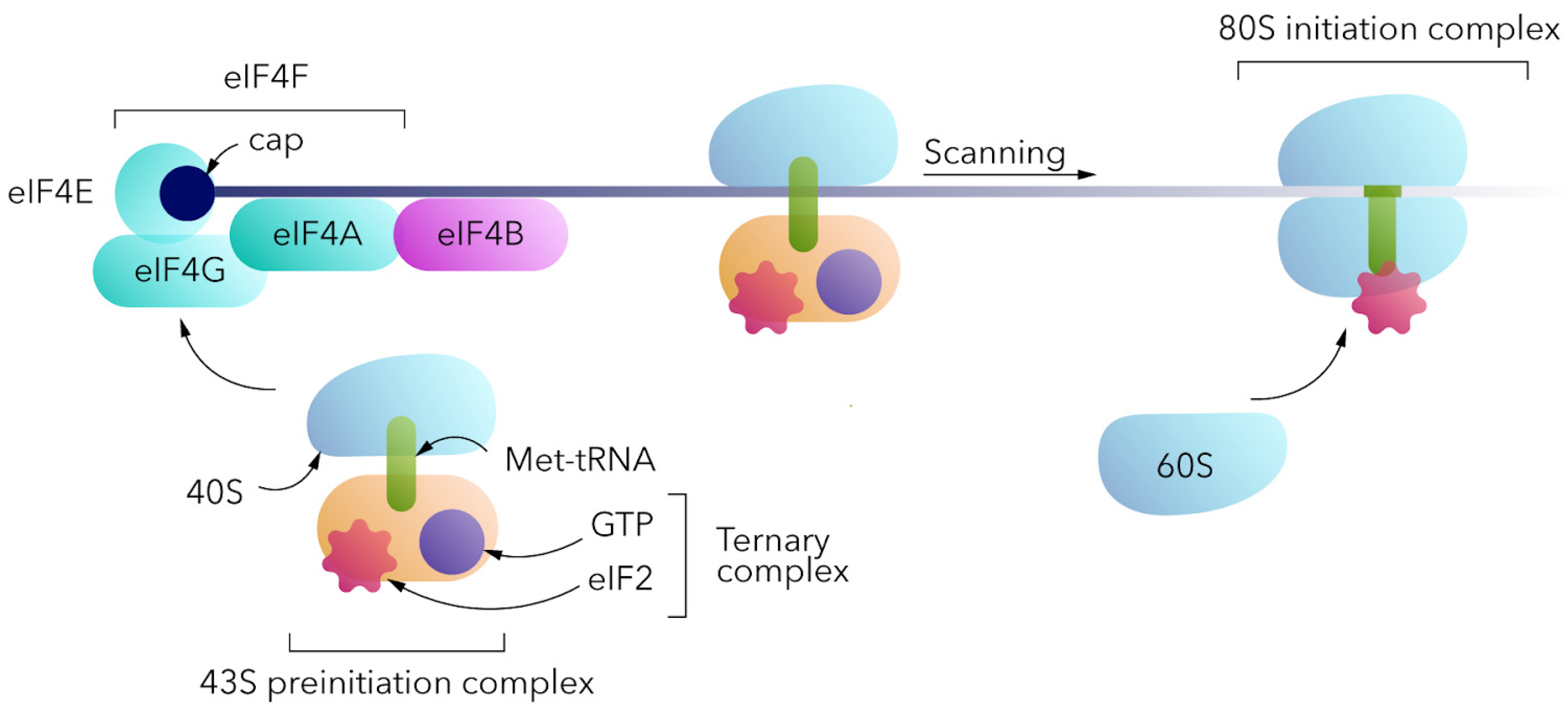
Figure 3. A simplified scheme of cap-dependent translation initiation mechanism.
2.1. Efficiency of IRES-Dependent Translation Initiation
The 5′UTR of mRNA has the potential to control translation independently of the cap via an internal ribosome entry site (IRES). Remarkably, cap-independent translation initiation can also be caused by other factors, which are discussed in the following sections. IRESs are present in both viral and cellular mRNAs. Cellular IRESs are very diverse, which makes them difficult to classify. In other words, there appear to be many different, as of yet uncharacterized, mechanisms that control cap-independent translation using endogenous IRESs [14].
Based on the differences in the way viral IRESs interact with ribosomes, they can be classified into four groups [15]. Group 1 IRESs bind to the ribosome without accessory factors. Group 2 IRESs bind to the ribosome with the help of eIFs and Met-tRNA. Group 3 IRESs bind to the ribosome using eIFs, Met-tRNA, as well as IRES transactivating factors (ITAFs), and function in rabbit reticulocyte lysate (RRL) without extracts from other cell types. Group 4 IRESs bind to the ribosome using eIFs, Met-tRNA, as well as ITAFs, and function in RRL only after addition of extracts from other cell types.
Originally, IRESs were studied as part of the viral replication system. Initial comparative studies showed low efficiency of IRES-associated translation initiation compared to cap-dependent initiation. In these studies, a bicitron system was used in which the first protein was synthesized in a classical cap-dependent manner and the second protein was translated by IRES. The efficiency of translation of the second protein was significantly lower [16,17][16][17]. The low protein expression induced by IRES was explained by the inefficient re-initiation of translation at an internal AUG codon downstream of IRES. IRES therefore gained a reputation as a less efficient translation initiation method as early as the 1980s, prompting researchers to modify IRES to improve the efficiency of translation initiation.
In the case of bicistronic cassettes, the efficiency of IRES-dependent translation initiation can be improved by a tailored intercistronic sequence [18]. The size of the intercistronic spacer sequence in the 5′ region of the IRES sequence has a significant impact on downstream protein expression. By choosing the spacer size, it was possible to achieve even stronger expression of proteins whose initiation was dependent on IRES and not on the cap. Another comparative study showed that IRES did not perform worse than cap under certain conditions [19]. Translation efficiency using EMCV IRES was similar compared to cap-dependent gene expression in the Lcl1D cell line, although efficiency remained low in many other cases [19]. This study compared the transcriptional efficiency of a gene in a bicistronic vector in which the initiation of the first protein was cap-dependent and that of the second protein was initiated by an EMCV IRES (type 2 IRES). Furthermore, it was shown that the efficiency of IRES strongly depends on the expressing cell line [19].
For more efficient use of IRES, it is worth choosing the correct combination of IRES type and express cell line. For example, group 1 IRESs (from PV, ECHO and HRV viruses) showed high translational efficiency in the HeLa, HepG2 and FRhK4 cell lines, while they were extremely inefficient in the BHK21 and Neuro-2A non-primate cell lines and in the SKNBE human neuronal cell line. At the same time, group 2 IRESs (from EMCV, FMDV and HCV) appear to be excellent candidates for the development of expression vectors, as they functioned efficiently in all cell lines tested by the authors [20].
Currently, there are several examples of successful developments based on IRES-mediated translation initiation. A circular mRNA prototype encoding the modified spike protein of SARS-CoV-2 induced neutralizing antibody titers for up to 7 weeks after secondary inoculation and had broad neutralizing activity against different viral strains [21]. In another study, circRNA was found to confer higher and more durable efficiency than analogous mRNA. The induced immunity persisted for at least 8 weeks after secondary inoculation [22].
2.2. m6A and Translation Initiation
The most common internal chemical RNA modification, N6-methyladenosine (m6A), is involved in the initiation of translation [28][23]. Even a single m6A modification within the 5′UTR of mRNA has the potential to recruit a 40S ribosomal subunit through direct eIF3 binding. Ribonucleotide methylation is a flexible process that depends on the activity of epitranscriptional “writer” and “eraser” factors [29][24].
The “readers” of the m6A modification bind mainly to a modified nucleotide in the RR(m6A)CH motif (R stands for purines, H for pyrimidines and C for the cytosine base). There are several representatives of the YTH domain-containing protein family: YTHDF1/2/3, localized in the cytoplasm of cells, and YTHDC1/2/3, localized in the nucleus [30][25].
Recently, a firefly luciferase assay was used to demonstrate that m6A-mediated translation initiation is less efficient compared to cap-dependent initiation [31][26]. On the other hand, this approach demonstrated comparable results in comparison to IRES-mediated translation initiation [32][27]. This technique can be applied in circular mRNA engineering, but the efficiency of such constructs should be carefully assessed.
2.3. Endogenous IRES-like Elements in Eukaryotic Genome
Several endogenous circRNAs are known to translate proteins in eukaryotic cells. Their 5′UTRs contain IRES-like sequences that drive translation independently of cap [33][28]. Fan et al. [23][29] found 97 short random hexamer sequences enriched in endogenous translating circRNAs (compared to linear mRNAs) that drive translation independently of cap. These IRES-like structures interact with various RNA-binding factors that initiate cap-independent protein expression. At the same time, RBPs are differentially expressed in various tissues [34][30]. This means that IRES-like sequences have different efficiencies in different tissues, which should be taken into consideration during implementation of IRES-like structures into genetic constructs. Mutation of these sequences significantly reduced the translation rate. In addition, some hexamers contain specific context-dependent m6A modifications of such RNAs, further complicating the system. Remarkably, virtually every random sequence longer than 50 nucleotides contains such a short IRES-like element. Indeed, the authors demonstrated that around 2% of all hexamers (97 out of 4096) may initiate translation. A sequence of 50 nucleotides can be divided into 45 possible hexamers, while a sequence of 100 nucleotides can be divided into 95 hexamers. Assuming that nucleotide sequences have some degree of randomness, wresearchers can conclude that any RNA sequence of any length can have translational potential if they contain an ORF. However, the identified elements showed great similarity with previously described IRES-like sequences of some endogenous genes, such as Hsp70 [35][31] and Gtx [36][32].
In addition, screenings have shown that almost 10% of eukaryotic mRNAs contain IRES-like sequences that can drive translation independently of the cap [37,38][33][34]. To date, it is not clear how effective such short sequences are in initiating translation compared to viral IRESs that have been previously used. At the same time, a comprehensive paper [23][29], published only half a year ago, showed that the efficiency of translation initiated by some hexamers is at least as good as initiation using the m6A modification.
2.4. Translation Enhancing Elements
Translation enhancing elements, TEE, are genomic sequences that facilitate cap-independent translation. They were first identified using a method based on the mRNA display principle [39][35]. InA this study, a genomic library consisting of approximately 1013 randomly selected 150-nucleotide fragments of total genomic DNA was created. These sequences were placed in the 5′-UTR of the starter DNA construct encoding a His6 affinity tag.
After in vitro transcription, the resulting set of uncapped single-stranded RNAs was conjugated by photoligation with a puromycin residue at the 3′-end and subjected to translation. Remarkably, during translation, a chemical bond was formed between the newly translated peptides and their coding mRNA by the natural peptidyl transferase activity of the ribosome, which recognized puromycin as a tyrosyl-tRNA analogue. As a result, molecules capable of cap-independent translation consisted of uncapped mRNAs linked to His-tags. In addition, these substances were selected by immobilized metal ion affinity chromatography. The functional RNAs were then isolated, reverse transcribed and amplified by PCR to create the novel pool of DNA for another selection cycle. This procedure was repeated six times until the sequences capable of cap-independent translation enhancement became predominant in the library. This approach revealed more than 12 thousand human genome elements mediating translation in a cap-independent manner in vitro and in mammalian cells.
Several TEEs have shown extremely high efficiency in protein production. For example, the addition of hTEE-658 to the capped construct increased the expression of the target protein 100-fold compared to a construct without TEE [40][36]. According to a further study, this 37-nucleotide motif placed upstream of the certain protein coding region of transfection plasmid was capable of increasing the protein expression level in experiments with transfection and subsequent infection using the vaccinia virus (VACV) [41][37]. Notably, neither the length of the 5′-leader sequence, nor the distance between the motif and the coding region affects the ability of hTEE-658 to increase transcription and translation rates. To the best of our knowledge, the mechanism of TEE cap-dependent translation enhancement or cap-independent translation initiation is unclear.
References
- Rodrigues, C.M.; Plotkin, S.A. Impact of vaccines; health, economic and social perspectives. Front. Microbiol. 2020, 11, 1526. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7371956/ (accessed on 17 January 2023).
- Crommelin, D.J.; Anchordoquy, T.J.; Volkin, D.B.; Jiskoot, W.; Mastrobattista, E. Addressing the Cold Reality of mRNA Vaccine Stability. J. Pharm. Sci. 2020, 110, 997–1001.
- Sharova, L.V.; Sharov, A.A.; Nedorezov, T.; Piao, Y.; Shaik, N.; Ko, M.S. Database for mRNA Half-Life of 19 977 Genes Obtained by DNA Microarray Analysis of Pluripotent and Differentiating Mouse Embryonic Stem Cells. DNA Res. 2009, 16, 45–58.
- Deutscher, M.P.; Li, Z. Exoribonucleases and their multiple roles in RNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 2000, 66, 67–105.
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984.
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2012, 19, 141–157.
- Chen, R.; Wang, S.K.; Belk, J.A.; Amaya, L.; Li, Z.; Cardenas, A.; Abe, B.T.; Chen, C.K.; Wender, P.A.; Chang, H.Y. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 2022, 40, 1–10.
- Meganck, R.M.; Liu, J.; Hale, A.E.; Simon, K.E.; Fanous, M.M.; Vincent, H.A.; Wilusz, J.E.; Moorman, N.J.; Marzluff, W.F.; Asokan, A. Engineering highly efficient backsplicing and translation of synthetic circRNAs. Mol. Ther. Nucleic Acids 2021, 23, 821–834.
- Wesselhoeft, R.A.; Kowalski, P.S.; Anderson, D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018, 9, 1–10.
- Lee, K.H.; Kim, S.; Lee, S.-W. Pros and Cons of In Vitro Methods for Circular RNA Preparation. Int. J. Mol. Sci. 2022, 23, 13247.
- Hinnebusch, A.G. Structural Insights into the Mechanism of Scanning and Start Codon Recognition in Eukaryotic Translation Initiation. Trends Biochem. Sci. 2017, 42, 589–611.
- Sonneveld, S.; Verhagen, B.M.; Tanenbaum, M.E. Heterogeneity in mRNA Translation. Trends Cell Biol. 2020, 30, 606–618.
- Eliseev, B.; Yeramala, L.; Leitner, A.; Karuppasamy, M.; Raimondeau, E.; Huard, K.; Alkalaeva, E.; Aebersold, R.; Schaffitzel, C. Structure of a human cap-dependent 48S translation pre-initiation complex. Nucleic Acids Res. 2018, 46, 2678–2689.
- Yang, Y.; Wang, Z. IRES-mediated cap-independent translation, a path leading to hidden proteome. J. Mol. Cell Biol. 2019, 11, 911–919.
- Kieft, J.S. Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 2008, 33, 274–283.
- Kozak, M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol. 1987, 7, 3438–3445.
- Peabody, D.S.; Berg, P. Termination-reinitiation occurs in the translation of mammalian cell mRNAs. Mol. Cell. Biol. 1986, 6, 2695–2703.
- Al-Allaf, F.A.; Abduljaleel, Z.; Athar, M.; Taher, M.M.; Khan, W.; Mehmet, H.; Colakogullari, M.; Apostolidou, S.; Bigger, B.; Waddington, S.; et al. Modifying inter-cistronic sequence significantly enhances IRES dependent second gene expression in bicistronic vector: Construction of optimised cassette for gene therapy of familial hypercholesterolemia. Non-Coding RNA Res. 2019, 4, 1–14.
- Mizuguchi, H.; Xu, Z.; Ishii-Watabe, A.; Uchida, E.; Hayakawa, T. IRES-Dependent Second Gene Expression Is Significantly Lower Than Cap-Dependent First Gene Expression in a Bicistronic Vector. Mol. Ther. 2000, 1, 376–382.
- Borman, A.M.; Le Mercier, P.; Girard, M.; Kean, K.M. Comparison of Picornaviral IRES-Driven Internal Initiation of Translation in Cultured Cells of Different Origins. Nucleic Acids Res. 1997, 25, 925–932.
- Seephetdee, C.; Bhukhai, K.; Buasri, N.; Leelukkanaveera, P.; Lerdwattanasombat, P.; Manopwisedjaroen, S.; Phueakphud, N.; Kuhaudomlarp, S.; Olmedillas, E.; Saphire, E.O.; et al. A circular mRNA vaccine prototype producing VFLIP-X spike confers a broad neutralization of SARS-CoV-2 variants by mouse sera. Antivir. Res. 2022, 204, 105370.
- Qu, L.; Yi, Z.; Shen, Y.; Lin, L.; Chen, F.; Xu, Y.; Wu, Z.; Tang, H.; Zhang, X.; Tian, F.; et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 2022, 185, 1728–1744.
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5′ UTR m6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010.
- Kostyusheva, A.; Brezgin, S.; Glebe, D.; Kostyushev, D.; Chulanov, V. Host-cell interactions in HBV infection and pathogenesis: The emerging role of m6A modification. Emerg. Microbes Infect. 2021, 10, 2264–2275.
- Yang, B.; Wang, J.Q.; Tan, Y.; Yuan, R.; Chen, Z.S.; Zou, C. RNA methylation and cancer treatment. Pharmacol. Res. 2021, 174, 105937.
- Sakharov, P.A.; Smolin, E.A.; Lyabin, D.N.; Agalarov, S.C. ATP-Independent Initiation during Cap-Independent Translation of m6A-Modified mRNA. Int. J. Mol. Sci. 2021, 22, 3662.
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N 6-methyladenosine. Cell Res. 2017, 27, 626–641.
- Sinha, T.; Panigrahi, C.; Das, D.; Chandra Panda, A. Circular RNA translation, a path to hidden proteome. Wiley Interdiscip. Rev. RNA 2021, 13, e1685.
- Fan, X.; Yang, Y.; Chen, C.; Wang, Z. Pervasive translation of circular RNAs driven by short IRES-like elements. Nat. Commun. 2022, 13, 1–15.
- Van Nostrand, E.L.; Freese, P.; Pratt, G.A.; Wang, X.; Wei, X.; Xiao, R.; Blue, S.M.; Chen, J.Y.; Cody, N.A.; Dominguez, D.; et al. A large-scale binding and functional map of human RNA-binding proteins. Nature 2020, 583, 711–719.
- Rubtsova, M.P.; Sizova, D.V.; Dmitriev, S.E.; Ivanov, D.S.; Prassolov, V.S.; Shatsky, I.N. Distinctive Properties of the 5′-Untranslated Region of Human Hsp70 mRNA. J. Biol. Chem. 2003, 278, 22350–22356.
- Chappell, S.A.; Edelman, G.M.; Mauro, V.P. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl. Acad. Sci. USA 2000, 97, 1536–1541.
- Spriggs, K.A.; Stoneley, M.; Bushell, M.; Willis, A.E. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol. Cell 2008, 100, 27–38.
- Weingarten-Gabbay, S.; Elias-Kirma, S.; Nir, R.; Gritsenko, A.A.; Stern-Ginossar, N.; Yakhini, Z.; Weinberger, A.; Segal, E. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 2016, 351, aad4939.
- Wellensiek, B.P.; Larsen, A.C.; Stephens, B.; Kukurba, K.; Waern, K.; Briones, N.; Liu, L.; Snyder, M.; Jacobs, B.L.; Kumar, S.; et al. Genome-wide profiling of human cap-independent translation-enhancing elements. Nat. Methods 2013, 10, 747–750.
- Wellensiek, B.P.; Larsen, A.C.; Flores, J.; Jacobs, B.L.; Chaput, J.C. A leader sequence capable of enhancing RNA expression and protein synthesis in mammalian cells. Protein Sci. 2013, 22, 1392–1398.
- Richard, H.B., Jr.; Minder, S.; Sidhu, A.; Juba, A.N.; Jancovich, J.K.; Jacobs, B.L.; Wellensiek, B.P. Optimization of translation enhancing element use to increase protein expression in a vaccinia virus system. J. Gen. Virol. 2021, 102, 001624.
More
