You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 3 by Jessie Wu and Version 4 by Jessie Wu.
Amyotrophic Lateral Sclerosis is a progressive neurodegenerative disease and is the most common adult motor neuron disease. The disease pathogenesis is complex with the perturbation of multiple pathways proposed, including mitochondrial dysfunction, RNA processing, glutamate excitotoxicity, endoplasmic reticulum stress, protein homeostasis and endosomal transport/extracellular vesicle (EV) secretion. EVs are nanoscopic membrane-bound particles that are released from cells, involved in the intercellular communication of proteins, lipids and genetic material, and there is increasing evidence of their role in amyotrophic lateral sclerosis (ALS).
- amyotrophic lateral sclerosis
- exosomes
- extracellular vesicles
1. Extracellular Vesicles in Amyotrophic Lateral Sclerosis
Extracellular vesicles (EVs) are secreted from almost all cells and circulate freely around the body. They are able to cross the blood–brain barrier and therefore are a means of intercellular signalling to and from the central nervous system (CNS) [1]. EVs contribute to many physiological processes in the CNS, including: neural growth and development; CNS inflammation; the neuroprotective response to oxidative stress; and maintaining brain vascular integrity and post synaptic retrograde signalling [2][3][4][5][6]. EVs are also implicated in the pathological processes of neurodegeneration, and large numbers of studies have in recent years evaluated the role of EVs in neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS), Parkinson’s Disease (PD), Alzheimer’s Disease (AD), Huntington’s Disease and prion diseases [7].
1.1. Amyotrophic Lateral Sclerosis Associated Genes Involved in Vesicular Pathways
There are now over 50 potentially causal or disease modifying genes identified for ALS [8]. The most commonly identified gene mutation in European and North American patients is the hexanucleotide repeat expansion in C9orf72, accounting for up to a third of all identified pathogenic mutations [9]. Other common causal mutations are in the genes SOD1, TARDBP, NEK1, FIG4 and TBK1 [9][10][11]. There is wide geographical variation in the genetic causes of ALS. Familial ALS is much less frequent in China (1.2–2.7% of total cases), where mutations in SOD1 are the most frequent genetic cause (25.3% of fALS), followed by TARDBP and FUS [12]. This is similar to Japan, where SOD1 mutations are the most common cause of fALS (29.8%), followed by FUS and TARDBP, whereas C9orf72 expansions are rare, identified in only 1.3% of fALS cases [13]. In Brazil, the most frequent genetic cause is VAPB (30% of all fALS) followed by C9orf72 (22%) [14].
The multiple genes involved in ALS have a range of effects on multiple cellular processes including RNA processing, protein homeostasis, cytoskeletal dynamics, mitochondrial function, endosomal trafficking, autophagy and, important for this research, the formation of EVs [8][15]. Multiple ALS associated genes are involved in vesicular trafficking and EV regulation as shown in Table 1. The CHMP2B protein is an essential component of the ESCRT III complex, which is part of the machinery for MVB and EV formation, as discussed above [16]. C9orf72, VAPB, FIG4, SPG11 and ALS2 encode proteins which are involved in intracellular trafficking of vesicles [17][18][19][20][21][22]. Optineurin and SQSTM1/p62 proteins are both autophagy receptors and are activated through phosphorylation by TBK1 [23]. Autophagy and EV function are closely linked, with the level of autophagy in cells closely related to the secretion and transport of EVs [24][25]. Table 1 lists some of the proposed pathogenic mechanisms for mutations of genes having functions related to vesicular pathways (also illustrated in Figure 1).
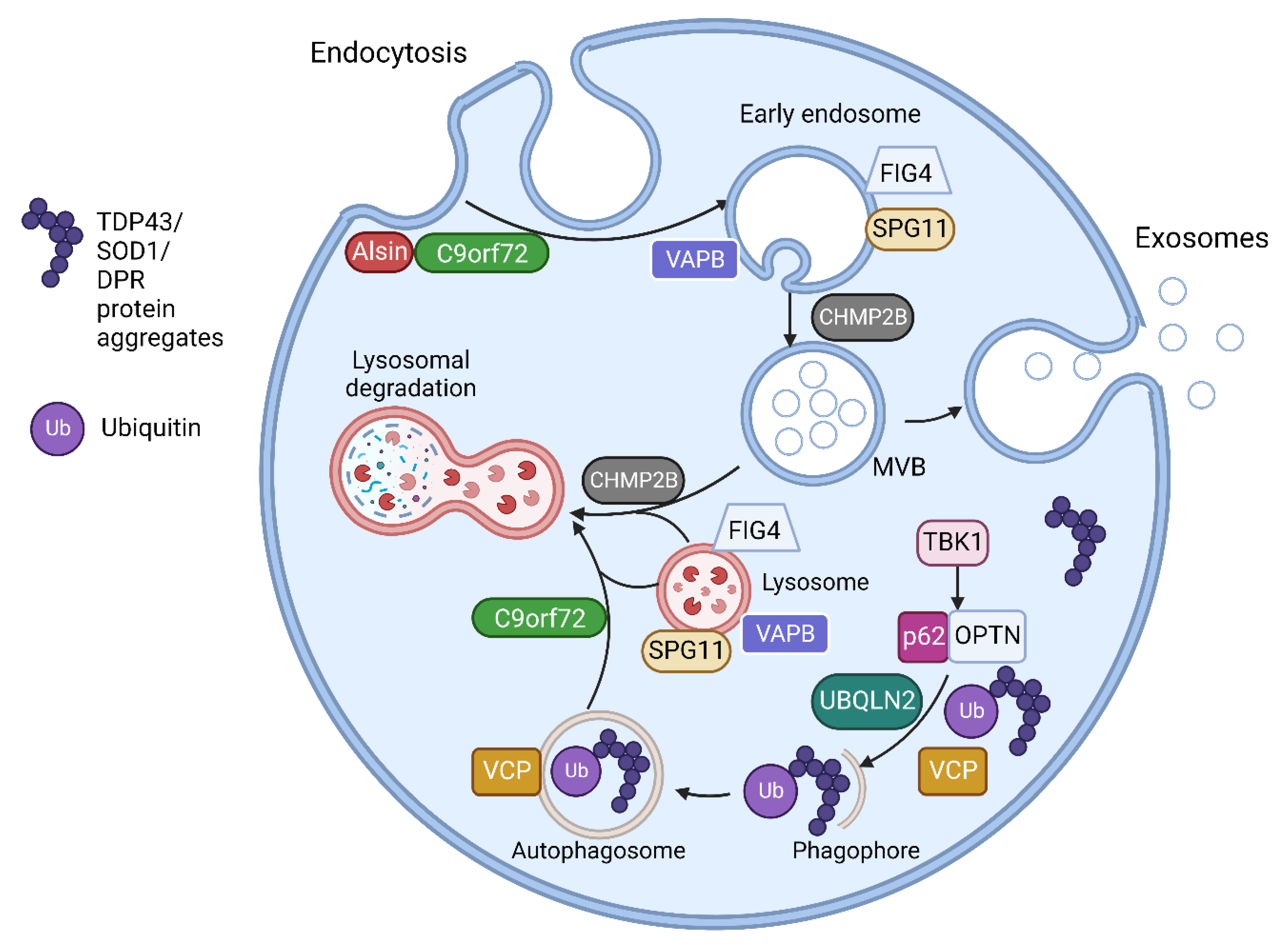
Figure 1. The endosomal and vesicular pathway involvement of proteins encoded by ALS related genes. Alsin activates Rab5 to promote endosomal fusion and subsequent endosomal trafficking. C9orf72 forms a complex with SMCR8 (Smith-Magenis Syndrome Chromosome Region, Candidate 8) and WDR41 (WD Repeat domain 41) proteins. This complex interacts with Rab GTPases including Rab5 in endosomal formation and trafficking. The C9orf72 complex also regulates various steps in autophagy including MVB-autophagosome and autophagosome-lysosome fusion as well as regulating several aspects of lysosomal biogenesis, pH and reformation. VAPB encodes Vesicle-associated membrane protein-associated protein B/C found in the endoplasmic reticulum (ER). This anchors complexes involved in lipid transfer from the ER to golgi and also the recycling of phosphatidylinositol-4-phosphate (PtdIns4P). VAPB mutations disrupt ER-golgi tethering and leads to PtdIns4P accumulation with subsequent accumulation of endosomes and dysfunctional lysosomes. FIG4 is required for the homeostasis of a signalling lipid phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2), which is required for endosomal and lysosomal maturation. FIG4 mutations result in enlarged endosomes and lysosomes with impaired lysosomal function. CHMP2B is responsible for the formation of intraluminal vesicles within the MVBs and may participate in the proper fusion of MVB with the lysosomes and the autophagosomes. CHMP2Bintron5 mutation results in accumulation of large endosomes and autophagosomes. VCP is involved in the initiation of autophagy and autophagosome maturation. TBK1 phosphorylates both optineurin (OPTN) and Sequestosome-1/p62, increasing their ability to bind to ubiquitinated cargo, initiating autophagy and delivery to autophagosomes. UBQLN2 interacts with LC3, a marker for starvation induced autophagy, to deliver ubiquitinated cargo to autophagosomes and is also recruited to OPTN containing vesicles. Spatacsin interacts with Rab5 for endosomal trafficking and maturation and contributes to lipid clearance from late endosomes and lysosomes. SPG11 mutations result in loss of spatacsin function, which leads to accumulation of lipids in lysosomes. Figure created using Biorender.com.
Table 1. ALS genes with roles in vesicular transport and EV regulation. Abbreviations: DPR—dipeptide repeat proteins, RBP—RNA binding proteins, UPS—ubiquitin proteasome system, NF-κB—nuclear factor kappa B.
| Gene | Proteins | Molecular Pathways Affected | ||
|---|---|---|---|---|
| C9orf72 | [17][26] | C9orf72 short and long isoforms | Loss of function in vesicle trafficking, autophagy and endo-lysosomal pathway Gain of toxicity with development of RNA foci and DPR |
|
| VAPB | [20][21] | Vesicle-associated membrane protein-associated protein B/C | Aggregation of VAPB protein, altered autophagy and vesicular transport, accumulation of RBPs | |
| FIG4 | [18] | Polyphosphoinositide phosphatase | Loss of function in trafficking of endosomal vesicles to golgi and autophagy regulation | |
| ALS2 | [19] | Alsin | Alteration of Rab5-mediated pathway with dysregulation of endosomal trafficking Altered trafficking of AMPA receptors causing glutamate toxicity |
|
| CHMP2B | [16] | Charged multivesicular body protein 2b |
Dysfunction of autophagy and endo-lysosomal pathway, resulting in accumulation of enlarged endosomes and autophagic organelles | |
| SPG11 | [22] | Spatacsin | Impaired autophagy, lipid sorting in late endosomes and lysosomal dysfunction with lipid accumulation | |
| SQSTM1 | [27] | Sequestosome-1/p62 | Dysfunction of autophagy and protein degradation through UPS | |
| OPTN | [28] | Optineurin | Golgi fragmentation, impaired autophagy and vesicular transport Loss of inhibitory action on NF-κB leading to abnormal inflammatory response |
|
| UBQLN2 | [29] | Ubiquilin 2 | Impaired protein degradation via UPS and dysfunction of autophagy and endo-lysosomal pathway | |
| VCP | [30][31] | Valosin Containing Protein | Impaired protein degradation via UPS and dysfunction of autophagy and endo-lysosomal pathway | |
| TBK1 | [32] | Tank Binding Kinase 1 | Dysregulation of multiple autophagy pathways |
1.2. Extracellular Vesicle Mediated Transfer of Misfolded Proteins and miRNAs in Amyotrophic Lateral LSclerosis
A pathological hallmark of ALS is TAR DNA-binding protein 43 (TDP 43) positive inclusions, which have been identified in brain stem and spinal cord tissue in over 97% of patients at postmortem [33]. SOD1, fused in sarcoma (FUS), and dipeptide repeat proteins (DPRs) from the C9orf72 intronic hexanucleotide repeat expansion, also aggregate to form protein inclusions [34]. These misfolded proteins have been demonstrated to spread between cells in a prion-like manner and induce further protein misfolding [35]. This mechanism has been used to explain the contiguous spread of disease that is often seen in ALS, where the disease spreads to adjacent neuroanatomical segments [36]. EVs have been shown to contain aberrant protein aggregates in cell and animal models of ALS as well as in patients with ALS. Evidence is growing to support the hypothesis that EVs can spread pathological misfolded proteins between cells in ALS and that these EVs can exert deleterious effects on recipient cells, discussed in Section 4 below. EVs have also been shown to mediate the transfer of RNAs, particularly miRNA, which can alter gene expression in recipient cells, potentially also of relevance to the progressive spread of neurodegeneration in ALS. Studies in other neurodegenerative diseases have also found that pathological proteins such as prion protein, amyloid-β, α-synuclein, and tau propagate via exosomes [37][38][39]. A timeline of discoveries in cell based and animal models of these roles of EVs in ALS is shown in Figure 2.
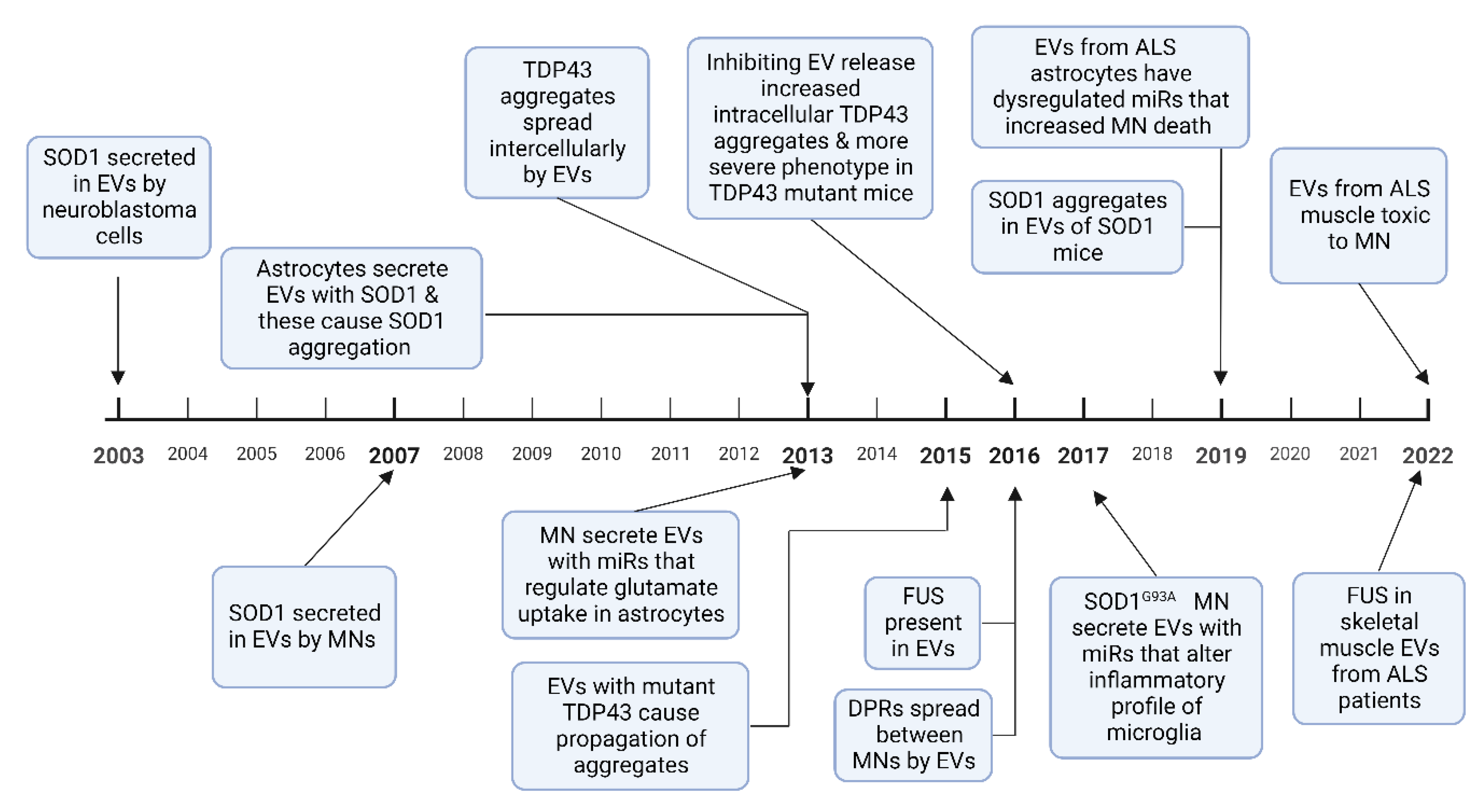
Figure 2. A timeline of experimental evidence of how EVs are involved in the spread of misfolded proteins, and how miRNA alter cellular phenotypes, in ALS cell-based and animal models. Abbreviations: MN-motor neuron, DPR-dipeptide repeat proteins [40][41][42][43][44][45][46][47][48][49][50][51][52]. Figure created using Biorender.com.
1.2.1. SOD1
The first evidence that EVs could spread misfolded proteins was in 2003 when SOD1 was shown to be excreted by EVs in SK-N-BE neuroblastoma cells [40]. Astrocytes with mutant SOD1 overexpression showed increased EV secretion compared to wild type cells; these secreted EVs were taken up by motor neurons, inducing cell death [42]. Misfolded SOD1 aggregates spread between NSC-34 motor neuron-like cells via EVs, causing cell rupture and cell death [41][53]. Once the SOD1 aggregates are introduced to neural cells, they result in a self-perpetuating induction of further SOD1 aggregation and transfer between cells [54]. These results have also been replicated in animal models, with CNS derived EVs in SOD1G93A mice containing misfolded SOD1 aggregates [50]. However, while mutant SOD1 aggregates are secreted by EVs, other studies have reported impaired secretion of mutant SOD1 compared to the wild type protein in NSC-34 motor neuron-like cells and rat microglial cells, with a proposed underlying mechanism of dysfunctional secretory pathways as a result of golgi fragmentation and ER stress [55][56].
1.2.2. TDP 43
Multiple studies demonstrate that TDP 43 is transferred intercellularly via EVs. Insoluble TDP 43 aggregates from ALS or FTD brain tissue resulted in intracellular accumulation of TDP 43 and cell death when added to SH-SY5Y neuroblastoma cells [43]. The same study also showed that TDP 43 aggregates can transfer between cells via EVs. CSF EVs in patients with ALS and ALS/FTD contain TDP 43 and these EVs have been shown to cause propagation of TDP 43 aggregates when added to U251 glioblastoma cells [45]. EVs isolated from human ALS brain tissue also caused TDP 43 accumulation and propagation in Neuro2a mouse glioblastoma cells [46]. In the same study, EV release was inhibited by a sphingomyelinase inhibitor, which resulted in increased TDP 43 aggregates in the Neuro2a cells and also exacerbated the clinical phenotype of transgenic mice expressing human mutant TDP-43A315T. This suggests that while EVs play a key role in the propagation of TDP 43 proteinopathy, the inhibition of EV secretion may precipitate greater intracellular accumulation of pathological aggregates [46]. Free TDP 43 can also be taken up by cells, but it has been shown in human embryonic kidney 293 (HEK-293) cells that EVs containing TDP 43 are preferentially taken up compared to free TDP 43, and thus have potential to spread and exert greater cellular toxicity [57].
1.2.3. FUS
There are few studies analysing mutant FUS or FUS binding partners. Analysis of HEK cells expressing mutant FUSR521G confirmed that FUS is present in EVs and that many FUS binding partners are components of EVs [47]. FUS and several FUS binding partners were also observed in EVs derived from skeletal muscle samples from sALS patients without FUS mutations [52].
1.2.4. Dipeptide Repeat Proteins
DPRs are a group of 5 protein complexes formed as a result of repeat-associated non-AUG (RAN) translation of the C9orf72 intronic hexanucleotide repeat expansion [58]. Such DPRs have been shown to spread intercellularly via EVs in spinal motor neurons derived from induced pluripotent stem cells (iPSCs) expressing the expanded hexanucleotide repeat [48].
1.2.5. RNA Transport by EVs
While multiple types of RNA have been found in EVs, most is known about miRNAs. These are short non-coding RNAs which have gained great attention due to their ability to modify gene expression in recipient cells [59]. MiRNAs are stable in the circulation and are found in a range of body fluids including serum, plasma, urine and CSF [60]. There is also evidence of their transport in EVs between cells [61]. EVs contain miRNA profiles that are vastly distinct from their host cell from which they originate [62]. Multiple miRNAs have been identified as dysregulated in vitro in ALS models involving several different cell types. EVs have been shown to transmit miR-124-3p from neurons to astrocytes, which regulates the glutamate uptake of astrocytes [63]. A study on astrocytes derived from iPSCs from C9orf72 ALS patients compared with iPSCs from healthy controls found that the EV miRNA content was dysregulated in ALS, with 64 dysregulated miRNAs, and downregulation of miR-494-3p as the most significant change [51].
There are fewer in vitro studies of messenger RNA (mRNA) expression in EVs in ALS. Similar to miRNA, mRNA expression in EVs from iPSC derived motor neurons is markedly different from that of the iPSC cells themselves, being enriched for genes regulating cellular metabolism and protein homeostasis [64].
2. Therapeutic Application of Extracellular Vesicles in Amyotrophic Lateral Sclerosis
There has been great interest in the potential use of EVs as a non-invasive method to deliver therapeutics including proteins, genetic material and drugs, in neurodegenerative diseases [65]. The investigation of EVs as therapeutic vectors is growing with at least 20 phase 1/2 clinical trials registered in patients with cancer, SARS-CoV-2, and Alzheimer’s disease, with treatments including stem cell derived EVs, autologous EVs or drug loaded EVs [66]. There are several properties of EVs which suggest that they could be useful in therapeutics in neurodegenerative diseases such as their ability to cross the blood–brain barrier, their low tendency to evoke an immune response and the potential for manipulation of cell surface markers to limit off target effects [67][68]. However, there are also challenges in development of EVs for this use, such as batch-to-batch variation in their synthesis, the current lack of cost-effective large scale production protocols or of robust, reproducible methods for drug loading [66]. EVs have been used in murine models of PD and AD to deliver small interfering RNAs (siRNA) to reduce pathological protein accumulation. EVs with α-Synuclein siRNA were peripherally injected into α-SynucleinS129D transgenic mice. This decreased the level of α-Synuclein aggregates in brain regions pathologically affected in PD [69]. EVs containing BACE1 siRNAs have also been used in C57BL/6 mice resulting in an over 60% reduction in BACE1 mRNA and a 55% decrease in β-amyloid 1-42 levels, a key component of plaques in AD pathology [70]. Studies in ALS have used EVs taken from mesenchymal stem cells (MSCs). The capacity of MSCs to replicate, differentiate, secrete neuroprotective factors and produce new cells to replace damaged cells, has led to multiple phase 1/2 clinical studies investigating their potential use in ALS [71]. However, challenges to the use of MSCs include dosing issues, variation in the differentiation state of the cells and the route of administration [72]. As it is now recognised that MSCs exert much of their action through secretion of EVs, investigation of the MSC secretome and EVs, as a cell-free therapeutic approach, is now being explored [73]. EVs from adipose derived stem cells (ADSCs) have been investigated in ALS cell models. Healthy human ADSCs were added to murine SOD1G93A NSC cells resulting in slower SOD1 aggregation and improved mitochondrial function [74]. EVs from Murine ADSCs have also been added to NSC-34 cells overexpressing SOD1G93A, SOD1G37R and SOD1A4V, which were challenged to oxidative damage with hydrogen peroxide. Treatment with EVs reduced oxidative damage, increased cell viability and improved mitochondrial function [75][76]. A subsequent study by the same group confirmed their previous findings and reported proteomic analysis on the ADSC EVs, identifying 189 proteins. Gene ontology analysis showed the most significant biological processes of the identified proteins to be cell adhesion, negative regulation of apoptosis and positive regulation of cell proliferation [77]. The group also tested intravenous and intranasal administration of ADSC EVs to SOD1G93A mice. The EVs were labelled with ultra-small superparamagnetic iron oxide nanoparticles and MRI confirmed the EVs did penetrate the blood–brain barrier and were deposited in the brain. The mice showed improved motor performance compared to placebo and had greater preservation of lumbar motor neurons, neuromuscular junctions and muscle fibres [78]. EVs isolated from human bone marrow-derived endothelial progenitor cells have also been shown to reduce damage in a murine brain endothelial cell ALS model [79]. Giunti et al. modified EVs from bone marrow derived MSCs by first treating the MSCs with interferon-γ. Treating the MSCs with interferon-γ resulted in upregulation of multiple microRNAs in EVs including miR-466q and miR-467f, which can reduce microglial activation through inhibition of the p38 mitogen-activated protein kinase pathway. The EVs reduced the levels of mRNA for Tumour Necrosis Factor and Interleukin 1b in SOD1G93A microglial cells [80]. This research showed that EV content can in principle be altered to exert the desired effect on target cells.References
- Banks, W.A.; Sharma, P.; Bullock, K.M.; Hansen, K.M.; Ludwig, N.; Whiteside, T.L. Transport of Extracellular Vesicles across the Blood-Brain Barrier: Brain Pharmacokinetics and Effects of Inflammation. Int. J. Mol. Sci. 2020, 21, 4407.
- Pascua-Maestro, R.; González, E.; Lillo, C.; Ganfornina, M.D.; Falcón-Pérez, J.M.; Sanchez, D. Extracellular Vesicles Secreted by As-troglial Cells Transport Apolipoprotein D to Neurons and Mediate Neuronal Survival Upon Oxidative Stress. Front. Cell Neurosci. 2019, 12, 526.
- Korkut, C.; Li, Y.; Koles, K.; Brewer, C.; Ashley, J.; Yoshihara, M.; Budnik, V. Regulation of Postsynaptic Retrograde Signaling by Presynaptic Exosome Release. Neuron 2013, 77, 1039–1046.
- Xu, B.; Zhang, Y.; Du, X.-F.; Li, J.; Zi, H.-X.; Bu, J.-W.; Yan, Y.; Han, H.; Du, J.-L. Neurons secrete miR-132-containing exosomes to regulate brain vascular integrity. Cell Res. 2017, 27, 882–897.
- Bahram Sangani, N.; Gomes, A.R.; Curfs, L.M.G.; Reutelingsperger, C.P. The role of Extracellular Vesicles during CNS development. Prog. Neurobiol. 2021, 205, 102124.
- Marostica, G.; Gelibter, S.; Gironi, M.; Nigro, A.; Furlan, R. Extracellular Vesicles in Neuroinflammation. Front. Cell Dev. Biol. 2021, 8, 623039.
- Thompson, A.G.; Gray, E.; Heman-Ackah, S.M.; Mäger, I.; Talbot, K.; El Andaloussi, S.; Wood, M.J.; Turner, M.R. Extracellular vesicles in neurodegenerative disease—Pathogenesis to biomarkers. Nat. Rev. Neurol. 2016, 12, 346–357.
- Mejzini, R.; Flynn, L.L.; Pitout, I.L.; Fletcher, S.; Wilton, S.D.; Akkari, P.A. ALS Genetics, Mechanisms, and Therapeutics: Where Are We Now? Front. Neurosci. 2019, 13, 1310.
- Grassano, M.; Calvo, A.; Moglia, C.; Sbaiz, L.; Brunetti, M.; Barberis, M.; Casale, F.; Manera, U.; Vasta, R.; Canosa, A.; et al. Systematic evaluation of genetic mutations in ALS: A population-based study. J. Neurol. Neurosurg. Psychiatry 2022, 93, 1190–1193.
- McCluskey, G.; Duddy, W.; Haffey, S.; Morrison, K.; Donaghy, C.; Duguez, S. Epidemiology and survival trends of motor neurone disease in Northern Ireland from 2015 to 2019. Eur. J. Neurol. 2021, 29, 707–714.
- Shepheard, S.R.; Parker, M.D.; Cooper-Knock, J.; Verber, N.S.; Tuddenham, L.; Heath, P.; Beauchamp, N.; Place, E.; Sollars, E.S.; Turner, M.R.; et al. Value of systematic genetic screening of patients with amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 2021, 92, 510–518.
- Liu, X.; He, J.; Gao, F.-B.; Gitler, A.D.; Fan, D. The epidemiology and genetics of Amyotrophic lateral sclerosis in China. Brain Res. 2018, 1693, 121–126.
- Suzuki, N.; Nishiyama, A.; Warita, H.; Aoki, M. Genetics of amyotrophic lateral sclerosis: Seeking therapeutic targets in the era of gene therapy. J. Hum. Genet. 2022, 2022, 1–22.
- Gonçalves, J.P.N.; Leoni, T.B.; Martins, M.P.; Peluzzo, T.M.; Dourado, M.E.T., Jr.; Saute, J.A.M.; Covaleski, A.P.P.M.; de Oliveira, A.S.B.; Claudino, R.; Marques, W., Jr.; et al. Genetic epidemiology of familial ALS in Brazil. Neurobiol. Aging 2021, 102, 227-e1–227-e4.
- Taylor, J.P.; Brown, R.H., Jr.; Cleveland, D.W. Decoding ALS: From genes to mechanism. Nature 2016, 539, 197–206.
- Ugbode, C.; West, R.J. Lessons learned from CHMP2B, implications for frontotemporal dementia and amyotrophic lateral sclerosis. Neurobiol. Dis. 2021, 147, 105144.
- Smeyers, J.; Banchi, E.G.; Latouche, M. C9ORF72: What It Is, What It Does, and Why It Matters. Front. Cell. Neurosci. 2021, 15, 661447.
- Chow, C.Y.; Landers, J.E.; Bergren, S.K.; Sapp, P.C.; Grant, A.E.; Jones, J.M.; Everett, L.; Lenk, G.M.; McKenna-Yasek, D.M.; Weisman, L.S.; et al. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am. J. Hum. Genet. 2009, 84, 85–88.
- Miceli, M.; Exertier, C.; Cavaglià, M.; Gugole, E.; Boccardo, M.; Casaluci, R.R.; Ceccarelli, N.; De Maio, A.; Vallone, B.; Deriu, M.A. ALS2-Related Motor Neuron Diseases: From Symptoms to Molecules. Biology 2022, 11, 77.
- Tripathi, P.; Guo, H.; Dreser, A.; Yamoah, A.; Sechi, A.; Jesse, C.M.; Katona, I.; Doukas, P.; Nikolin, S.; Ernst, S.; et al. Pathomechanisms of ALS8: Altered autophagy and defective RNA binding protein (RBP) homeostasis due to the VAPB P56S mutation. Cell Death Dis. 2021, 12, 466.
- Mao, D.; Lin, G.; Tepe, B.; Zuo, Z.; Tan, K.L.; Senturk, M.; Zhang, S.; Arenkiel, B.R.; Sardiello, M.; Bellen, H.J. VAMP associated proteins are required for autophagic and lysosomal degradation by promoting a PtdIns4P-mediated endosomal pathway. Autophagy 2019, 15, 1214–1233.
- Branchu, J.; Boutry, M.; Sourd, L.; Depp, M.; Leone, C.; Corriger, A.; Vallucci, M.; Esteves, T.; Matusiak, R.; Dumont, M.; et al. Loss of spatacsin function alters lysosomal lipid clearance leading to upper and lower motor neuron degeneration. Neurobiol. Dis. 2017, 102, 21–37.
- Richter, B.; Sliter, D.A.; Herhaus, L.; Stolz, A.; Wang, C.; Beli, P.; Zaffagnini, G.; Wild, P.; Martens, S.; Wagner, S.A.; et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl. Acad. Sci. 2016, 113, 4039–4044.
- Zheng, J.; Tan, J.; Miao, Y.-Y.; Zhang, Q. Extracellular vesicles degradation pathway based autophagy lysosome pathway. Am. J. Transl. Res. 2019, 11, 1170–1183.
- Wei, W.; Pan, Y.; Yang, X.; Chen, Z.; Heng, Y.; Yang, B.; Pu, M.; Zuo, J.; Lai, Z.; Tang, Y.; et al. The Emerging Role of the Interaction of Extracellular Vesicle and Au-tophagy-Novel Insights into Neurological Disorders. J. Inflamm. Res. 2022, 15, 3395–3407.
- Pang, W.; Hu, F. Cellular and physiological functions of C9ORF72 and implications for ALS/FTD. J. Neurochem. 2020, 157, 334–350.
- Davidson, J.M.; Chung, R.S.; Lee, A. The converging roles of sequestosome-1/p62 in the molecular pathways of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Neurobiol. Dis. 2022, 166, 105653.
- Markovinovic, A.; Cimbro, R.; Ljutic, T.; Kriz, J.; Rogelj, B.; Munitic, I. Optineurin in amyotrophic lateral sclerosis: Multifunctional adaptor protein at the crossroads of different neuroprotective mechanisms. Prog. Neurobiol. 2017, 154, 1–20.
- Osaka, M.; Ito, D.; Yagi, T.; Nihei, Y.; Suzuki, N. Evidence of a link between ubiquilin 2 and optineurin in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2014, 24, 1617–1629.
- Scarian, E.; Fiamingo, G.; Diamanti, L.; Palmieri, I.; Gagliardi, S.; Pansarasa, O. The Role of VCP Mutations in the Spectrum of Am-yotrophic Lateral Sclerosis-Frontotemporal Dementia. Front. Neurol. 2022, 13, 841394.
- Ferrari, V.; Cristofani, R.; Tedesco, B.; Crippa, V.; Chierichetti, M.; Casarotto, E.; Cozzi, M.; Mina, F.; Piccolella, M.; Galbiati, M.; et al. Valosin Containing Protein (VCP): A Multistep Regulator of Autophagy. Int. J. Mol. Sci. 2022, 23, 1939.
- Oakes, J.A.; Davies, M.C.; Collins, M.O. TBK1: A new player in ALS linking autophagy and neuroinflammation. Mol. Brain 2017, 10, 5.
- Mackenzie, I.R.; Bigio, E.H.; Ince, P.G.; Geser, F.; Neumann, M.; Cairns, N.J.; Kwong, L.K.; Forman, M.S.; Ravits, J.; Stewart, H.; et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann. Neurol. 2007, 61, 427–434.
- Cicardi, M.E.; Marrone, L.; Azzouz, M.; Trotti, D. Proteostatic imbalance and protein spreading in amyotrophic lateral sclerosis. EMBO J. 2021, 40, e106389.
- McAlary, L.; Plotkin, S.S.; Yerbury, J.J.; Cashman, N.R. Prion-Like Propagation of Protein Misfolding and Aggregation in Amyo-trophic Lateral Sclerosis. Front. Mol. Neurosci. 2019, 12, 262.
- Kanouchi, T.; Ohkubo, T.; Yokota, T. Can regional spreading of amyotrophic lateral sclerosis motor symptoms be explained by prion-like propagation? J. Neurol. Neurosurg. Psychiatry 2012, 83, 739–745.
- Hartmann, A.; Muth, C.; Dabrowski, O.; Krasemann, S.; Glatzel, M. Exosomes and the Prion Protein: More than One Truth. Front. Neurosci. 2017, 11, 194.
- Weng, S.; Lai, Q.-L.; Wang, J.; Zhuang, L.; Cheng, L.; Mo, Y.; Liu, L.; Zhao, Z.; Zhang, Y.; Qiao, S. The Role of Exosomes as Mediators of Neuroinflammation in the Pathogenesis and Treatment of Alzheimer’s Disease. Front. Aging Neurosci. 2022, 14, 899944.
- Tsunemi, T.; Ishiguro, Y.; Yoroisaka, A.; Hattori, N. Analysis of α-Synuclein in Exosomes. Methods Mol. Biol. 2021, 2322, 41–45.
- Mondola, P.; Ruggiero, G.; Serù, R.; Damiano, S.; Grimaldi, S.; Garbi, C.; Monda, M.; Greco, D.; Santillo, M. The Cu,Zn superoxide dismutase in neuroblastoma SK-N-BE cells is exported by a microvesicles dependent pathway. Mol. Brain Res. 2003, 110, 45–51.
- Gomes, C.; Keller, S.; Altevogt, P.; Costa, J. Evidence for secretion of Cu,Zn superoxide dismutase via exosomes from a cell model of amyotrophic lateral sclerosis. Neurosci. Lett. 2007, 428, 43–46.
- Basso, M.; Pozzi, S.; Tortarolo, M.; Fiordaliso, F.; Bisighini, C.; Pasetto, L.; Spaltro, G.; Lidonnici, D.; Gensano, F.; Battaglia, E.; et al. Mutant copper-zinc superoxide dismutase (SOD1) induces protein secretion pathway alterations and exosome release in astrocytes: Implications for disease spreading and motor neuron pathology in amyotrophic lateral sclerosis. J. Biol. Chem. 2013, 288, 15699–15711.
- Nonaka, T.; Masuda-Suzukake, M.; Arai, T.; Hasegawa, Y.; Akatsu, H.; Obi, T.; Yoshida, M.; Murayama, S.; Mann, D.M.; Akiyama, H.; et al. Prion-like Properties of Pathological TDP-43 Aggregates from Diseased Brains. Cell Rep. 2013, 4, 124–134.
- Morel, L.; Regan, M.; Higashimori, H.; Ng, S.K.; Esau, C.; Vidensky, S.; Rothstein, J.; Yang, Y. Neuronal exosomal miRNA-dependent translational regulation of astroglial glutamate transporter GLT1. J. Biol. Chem. 2013, 288, 7105–7116.
- Ding, X.; Ma, M.; Teng, J.; Teng, R.K.; Zhou, S.; Yin, J.; Fonkem, E.; Huang, J.H.; Wu, E.; Wang, X. Exposure to ALS-FTD-CSF generates TDP-43 aggregates in glioblastoma cells through exosomes and TNTs-like structure. Oncotarget 2015, 6, 24178–24191.
- Iguchi, Y.; Eid, L.; Parent, M.; Soucy, G.; Bareil, C.; Riku, Y.; Kawai, K.; Takagi, S.; Yoshida, M.; Katsuno, M.; et al. Exosome secretion is a key pathway for clearance of pathological TDP-43. Brain 2016, 139, 3187–3201.
- Kamelgarn, M.; Chen, J.; Kuang, L.; Arenas, A.; Zhai, J.; Zhu, H.; Gal, J. Proteomic analysis of FUS interacting proteins provides insights into FUS function and its role in ALS. Biochim. Biophys. Acta BBA-Mol. Basis Dis. 2016, 1862, 2004–2014.
- Westergard, T.; Jensen, B.K.; Wen, X.; Cai, J.; Kropf, E.; Iacovitti, L.; Pasinelli, P.; Trotti, D. Cell-to-Cell Transmission of Dipeptide Repeat Proteins Linked to C9orf72 -ALS/FTD. Cell Rep. 2016, 17, 645–652.
- Pinto, S.; Cunha, C.; Barbosa, M.; Vaz, A.R.; Brites, D. Exosomes from NSC-34 Cells Transfected with hSOD1-G93A Are Enriched in miR-124 and Drive Alterations in Microglia Phenotype. Front. Neurosci. 2017, 11, 273.
- Silverman, J.M.; Christy, D.; Shyu, C.C.; Moon, K.-M.; Fernando, S.; Gidden, Z.; Cowan, C.M.; Ban, Y.; Stacey, R.G.; Grad, L.I.; et al. CNS-derived extracellular vesicles from superoxide dismutase 1 (SOD1)G93A ALS mice originate from astrocytes and neurons and carry misfolded SOD1. J. Biol. Chem. 2019, 294, 3744–3759.
- Varcianna, A.; Myszczynska, M.A.; Castelli, L.M.; O’Neill, B.; Kim, Y.; Talbot, J.; Nyberg, S.; Nyamali, I.; Heath, P.R.; Stopford, M.J.; et al. Micro-RNAs secreted through astrocyte-derived extracellular vesicles cause neuronal network degeneration in C9orf72 ALS. Ebiomedicine 2019, 40, 626–635.
- Le Gall, L.; Duddy, W.J.; Martinat, C.; Mariot, V.; Connolly, O.; Milla, V.; Anakor, E.; Ouandaogo, Z.G.; Millecamps, S.; Lainé, J.; et al. Muscle cells of sporadic amyotrophic lateral sclerosis patients secrete neurotoxic vesicles. J. Cachexia Sarcopenia Muscle 2022, 13, 1385–1402.
- Grad, L.I.; Yerbury, J.J.; Turner, B.J.; Guest, W.C.; Pokrishevsky, E.; O’Neill, M.A.; Yanai, A.; Silverman, J.M.; Zeineddine, R.; Corcoran, L.; et al. Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. USA 2014, 111, 3620–3625.
- Münch, C.; O’Brien, J.; Bertolotti, A. Prion-like propagation of mutant superoxide dismutase-1 misfolding in neuronal cells. Proc. Natl. Acad. Sci. 2011, 108, 3548–3553.
- Massenzio, F.; Peña-Altamira, E.; Petralla, S.; Virgili, M.; Zuccheri, G.; Miti, A.; Polazzi, E.; Mengoni, I.; Piffaretti, D.; Monti, B. Microglial overexpression of fALS-linked mutant SOD1 induces SOD1 processing impairment, activation and neurotoxicity and is counteracted by the autophagy inducer trehalose. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 3771–3785.
- Turner, B.J.; Atkin, J.; Farg, M.A.; Zang, D.W.; Rembach, A.; Lopes, E.C.; Patch, J.D.; Hill, A.; Cheema, S.S. Impaired Extracellular Secretion of Mutant Superoxide Dismutase 1 Associates with Neurotoxicity in Familial Amyotrophic Lateral Sclerosis. J. Neurosci. 2005, 25, 108–117.
- Feiler, M.S.; Strobel, B.; Freischmidt, A.; Helferich, A.M.; Kappel, J.; Brewer, B.M.; Li, D.; Thal, D.; Walther, P.; Ludolph, A.C.; et al. TDP-43 is intercellularly transmitted across axon terminals. J. Cell Biol. 2015, 211, 897–911.
- Schmitz, A.; Marques, J.P.; Oertig, I.; Maharjan, N.; Saxena, S. Emerging Perspectives on Dipeptide Repeat Proteins in C9ORF72 ALS/FTD. Front. Cell. Neurosci. 2021, 15, 637548.
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606.
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010, 56, 1733–1741.
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659.
- Jovičić, A.; Gitler, A.D. Distinct repertoires of microRNAs present in mouse astrocytes compared to astrocyte-secreted exosomes. PLoS ONE 2017, 12, e0171418.
- Men, Y.; Yelick, J.; Jin, S.; Tian, Y.; Chiang, M.S.R.; Higashimori, H.; Brown, E.; Jarvis, R.; Yang, Y. Exosome reporter mice reveal the involvement of exosomes in mediating neuron to astroglia communication in the CNS. Nat. Commun. 2019, 10, 1–18.
- Otake, K.; Adachi-Tominari, K.; Nagai, H.; Saito, M.; Sano, O.; Hirozane, Y.; Iwata, H. Quantitative comparison of the mRNA content of human iPSC-derived motor neurons and their extracellular vesicles. FEBS Open Bio 2021, 11, 494–506.
- Garbuzova-Davis, S.; Sadanandan, N.; Lee, J.-Y. Extracellular vesicle-based therapy for amyotrophic lateral sclerosis. Brain Circ. 2021, 7, 23–28.
- Herrmann, I.K.; Wood, M.J.A.; Fuhrmann, G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 2021, 16, 748–759.
- Gagliardi, D.; Bresolin, N.; Comi, G.P.; Corti, S. Extracellular vesicles and amyotrophic lateral sclerosis: From misfolded protein vehicles to promising clinical biomarkers. Cell. Mol. Life Sci. 2020, 78, 561–572.
- Kalani, A.; Tyagi, N. Exosomes in neurological disease, neuroprotection, repair and therapeutics: Problems and perspectives. Neural Regen. Res. 2015, 10, 1565–1567.
- Cooper, J.M.; Wiklander, P.B.O.; Nordin, J.Z.; Al-Shawi, R.; Wood, M.J.; Vithlani, M.; Schapira, A.H.V.; Simons, J.P.; El-Andaloussi, S.; Alvarez-Erviti, L. Systemic exosomal siRNA delivery reduced alpha-synuclein aggregates in brains of transgenic mice. Mov. Disord. 2014, 29, 1476–1485.
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J.A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345.
- Gugliandolo, A.; Bramanti, P.; Mazzon, E. Mesenchymal Stem Cells: A Potential Therapeutic Approach for Amyotrophic Lateral Sclerosis? Stem Cells Int. 2019, 2019, 3675627.
- Ciervo, Y.; Ning, K.; Jun, X.; Shaw, P.J.; Mead, R.J. Advances, challenges and future directions for stem cell therapy in amyotrophic lateral sclerosis. Mol. Neurodegener. 2017, 12, 85.
- Araldi, R.P.; D’Amelio, F.; Vigerelli, H.; De Melo, T.C.; Kerkis, I. Stem Cell-Derived Exosomes as Therapeutic Approach for Neurodegenerative Disorders: From Biology to Biotechnology. Cells 2020, 9, 2663.
- Lee, M.; Ban, J.-J.; Kim, K.Y.; Jeon, G.S.; Im, W.; Sung, J.-J.; Kim, M. Adipose-derived stem cell exosomes alleviate pathology of amyotrophic lateral sclerosis in vitro. Biochem. Biophys. Res. Commun. 2016, 479, 434–439.
- Bonafede, R.; Scambi, I.; Peroni, D.; Potrich, V.; Boschi, F.; Benati, D.; Bonetti, B.; Mariotti, R. Exosome derived from murine adipose-derived stromal cells: Neuroprotective effect on in vitro model of amyotrophic lateral sclerosis. Exp. Cell Res. 2016, 340, 150–158.
- Calabria, E.; Scambi, I.; Bonafede, R.; Schiaffino, L.; Peroni, D.; Potrich, V.; Capelli, C.; Schena, F.; Mariotti, R. ASCs-Exosomes Recover Coupling Efficiency and Mitochondrial Membrane Potential in an in vitro Model of ALS. Front. Neurosci. 2019, 13, 1070.
- Bonafede, R.; Brandi, J.; Manfredi, M.; Scambi, I.; Schiaffino, L.; Merigo, F.; Turano, E.; Bonetti, B.; Marengo, E.; Cecconi, D.; et al. The Anti-Apoptotic Effect of ASC-Exosomes in an In Vitro ALS Model and Their Proteomic Analysis. Cells 2019, 8, 1087.
- Bonafede, R.; Turano, E.; Scambi, I.; Busato, A.; Bontempi, P.; Virla, F.; Schiaffino, L.; Marzola, P.; Bonetti, B.; Mariotti, R. ASC-Exosomes Ameliorate the Disease Progression in SOD1(G93A) Murine Model Underlining Their Potential Therapeutic Use in Human ALS. Int. J. Mol. Sci. 2020, 21, 3651.
- Garbuzova-Davis, S.; Willing, A.E.; Ehrhart, J.; Wang, L.; Sanberg, P.R.; Borlongan, C.V. Cell-Free Extracellular Vesicles Derived from Human Bone Marrow Endothelial Progenitor Cells as Potential Therapeutics for Microvascular Endothelium Restoration in ALS. NeuroMolecular Med. 2020, 22, 503–516.
- Giunti, D.; Marini, C.; Parodi, B.; Usai, C.; Milanese, M.; Bonanno, G.; de Rosbo, N.K.; Uccelli, A. Role of miRNAs shuttled by mesenchymal stem cell-derived small extracellular vesicles in modulating neuroinflammation. Sci. Rep. 2021, 11, 1740.
More
