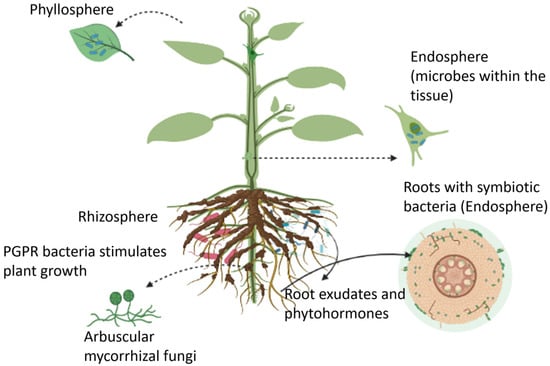
Plant-microbe interactions are critical for ecosystem functioning and driving rhizosphere processes. To fully understand the communication pathways between plants and rhizosphere microbes, it is crucial to measure the numerous processes that occur in the plant and the rhizosphere. Plants can host a wide range of microbes, collectively known as the plant microbiome, in the rhizosphere (i.e., the region of soil in the vicinity of plant roots), endosphere (i.e., plant internal tissues), and phyllosphere (i.e., stem, leaves, or flowers). These microbiomes form long-lasting interactions with the host plant, leading to positive, neutral, or negative impacts on crop performance and microbe-mediated biogeochemical processes.
1. Introduction
Microbial diversity exists in a natural soil environment with up to 10
10 microbial cells along with tens of thousands of bacteria and archaea living under the surface of each gram of soil
[1]. Some of these microbes, such as mycorrhizal fungi and nitrogen-fixing symbiotic bacteria substantially contribute to plant nutrition through recycling and utilizing the soil organic carbon as a source of energy
[2], fertilizing crops by providing nutrients, controlling or inhibiting plant pathogens, enhancing soil structure by forming microaggregates, mineralizing the organic pollutants in soil, and reducing the reliance on chemical fertilizers to achieve high productivity
[3][4][3,4]. These free-living soil bacteria are collectively called plant growth-promoting rhizobacteria (PGPR) that invade the root system and facilitate plant development by several mechanisms
[5][6][5,6]. They are also known as plant health-promoting rhizobacteria (PHPR) or nodule-promoting rhizobacteria (NPR) that stimulate plant-microbe interactions in the rhizosphere, a crucial soil ecosystem habitat
[5]. The biogeochemical processes in the rhizosphere influence the activity and composition of the plant’s microbial community. Increasing evidence suggests that bacteria such as Pseudomonas, Azospirillum, Azotobacter, Klebsiella, Enterobacter, Alcaligenes, Arthrobacter, Burkholderia, Bacillus, and Serratia promote plant growth and thus serve as PGPR
[7].
Plant-microbe interactions are mutual. The rhizosphere, a complex zone surrounding plant roots, is influenced by root secretions, plant species and their developmental stages, soil properties, nutrient status, land use, and climatic conditions
[8]. Studies suggest that the host plant’s unique composition of root exudates plays a major role in determining how the plant’s microbiome is structured, indicating the selective effect of a host plant on plant-microbe interactions
[1][9][1,9]. These exudates account for 5–21% of total photosynthetically fixed carbon-containing signaling and chemoattractant molecules. These molecules help in recruiting beneficial microorganisms that contribute to pathogen resistance, water retention, and the synthesis of growth-promoting hormones that influence plant phenotype
[10]. There are also harmful microbes, termed pathogens, which cause damage to the host plant. Plants recognize and respond to these pathogenic infections via the expression of specific defense or signaling molecules called phytohormones
[11]. A better understanding of rhizosphere microbiota and plant health would help manipulate the soil microbiome directly by incorporating specific microbes in the soil or indirectly by modifying management practices to improve crop performance
[8]. Moreover, understanding the dynamics and crosstalk between the hormonal signaling pathways would elucidate the defense mechanisms in plants
[12].
An analysis of the host plant together with its associated microbiome, typically called holobiont, suggests the coevolution of plants and microbes
[13]. Many modern technologies such as next-generation sequencing (NGS)
[14], computational tools, omics approaches (metagenomics, transcriptomics, proteomics, metabolomics), and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-based tools have revealed promise for understanding the molecular aspects of plant-microbe interactions, which underlie sustainable agricultural practices
[15]. As an analytical tool, biosensors have been extensively used to detect multifarious target substrates in the last few decades. These biosensors convert the chemical interactions into a measurable optical, electrical, or acoustic response
[16]. A biosensor is expected to detect target molecules with a high signal-to-noise ratio (SNR), provide high spatial and temporal resolution at the cellular/molecular level, respond quickly, and work under varying environmental conditions such as changes in temperature, pH, or redox states. In addition, the detection procedure should not interfere with cellular processes, cause cellular damage or incur any toxicity
[17]. There is a wide variety of biosensors reported in the literature
[18]. Over the past ten years, many studies have been published on plant biosensors, demonstrating the progress of this technology and its significance in plant research. One group is pursuing wearable, flexible, and wireless sensor design for in situ monitoring of phytohormone signaling and dynamics in plants
[19][20][21][22][23][19,20,21,22,23].
2. The Interplay between Plants and Microbial Communities
Plants can host a wide range of microbes, collectively known as the plant microbiome, in the rhizosphere (i.e., the region of soil in the vicinity of plant roots), endosphere (i.e., plant internal tissues), and phyllosphere (i.e., stem, leaves, or flowers) as illustrated in
Figure 1 [24]. These microbiomes form long-lasting interactions with the host plant, leading to positive, neutral, or negative impacts on crop performance and microbe-mediated biogeochemical processes
[25]. The microbial community that is beneficial to the host plant’s health, function, and evolution, are termed microsymbionts for forming a symbiotic relationship with the plant. The other type of microbe functions as plant pathogens, causing damage to the host plant. In most cases, the beneficial effects of microorganisms on plants are not caused by a single microbe, but rather by a consortium of different microorganisms that induce systemic resistance and promote plant growth
[26]. Berendsen et al., reported the prevalence of three bacterial genera,
Microbacterium,
Stenotrophomonas, and
Xanthomonas, in the rhizosphere of
Arabidopsis thaliana when the foliar defense was activated by the downy mildew pathogen
Hyaloperonospora arabidopsidis [27]. This study revealed that the plant recruited these bacterial species in the root zone to induce systemic resistance against downy mildew. Moreover, the formation of this symbiotic relationship in the primary population of downy mildew-infected plants resulted in a higher chance of survival of the second population of plants grown in the same soil. Microbiomes found in the endosphere and rhizosphere regions have also been shown to suppress plant diseases caused by fungal pathogens
Gaeumannomyces graminis and
Rhizoctonia solani (a soil-born pathogen)
[28][29][28,29]. Similarly, other studies suggested the impact of a consortium of endophytes, including the fungi
Rhodotorula graminis, and the bacteria
Burkholderia vietnamiensis,
Rahnella sp.,
Burkholderia sp.,
Acinetobacter calcoaceticus,
Sphingomonas yanoikuyae,
Pseudomonas sp.,
Rhizobium tropici, and
Curtobacterium sp. on the enhanced drought stress tolerance in poplar plants
[30]. In another study, pepper plants inoculated with desert-adapted bacteria displayed higher tolerance to water shortage compared with control plants. The bacteria enhanced the root biomass and length of plants (by 40%), which in turn improved the plant’s ability to uptake water and survive under water stress conditions
[31]. Furthermore, mutualism between plants and microbes increases nutrient availability for plants. Beneficial interactions of the host plant with the microbial community contribute to the co-existence of multiple plant species, thereby enhancing plant and microbial diversity. The heterospecific plant-soil feedback responses play an important role in the co-existence of species, ecological succession, and species invasiveness. A meta-analysis conducted by Kutakova et al., suggest that plants grew better in soil conditioned by their closely related species than in soil conditioned by less frequently co-occurring species
[32].
Figure 1.
An overview of plant growth-promoting microbes that reside in the rhizosphere, endosphere, and phyllosphere regions.
However, microbes that act as plant pathogens can directly infect the seedlings and suppress beneficial interactions. There are three main categories of pathogens:
biotrophs, that feed on nutrients while keeping the host plant alive;
necrotrophs, that suppress and destabilize the host’s immune system by producing tissue-degrading toxins and enzymes and feed on the dead tissue; and
hemibiotrophs, that initially behave like
biotrophs but the transition to
necrotrophs in later stages of the disease
[11]. For instance,
Pseudomonas syringae (
P. syringae) strains secrete the effector molecule, AvrPto1, which suppresses immune-related proteins in tomato plants. These effector molecules are specific to the pathogen
[33].
The plant produces signaling hormones or phytohormones that include salicylic acid (SA), jasmonic acid (JA), ethylene (ET), abscisic acid (ABA), and auxins (such as indole-3-acetic acid (IAA)). The phytohormone signals are generated in the infected tissue and then circulated throughout the plant via the xylem and phloem. A progressive variation in the phytohormone levels serves as an early signal of plant stress
[34][35][36][34,35,36].
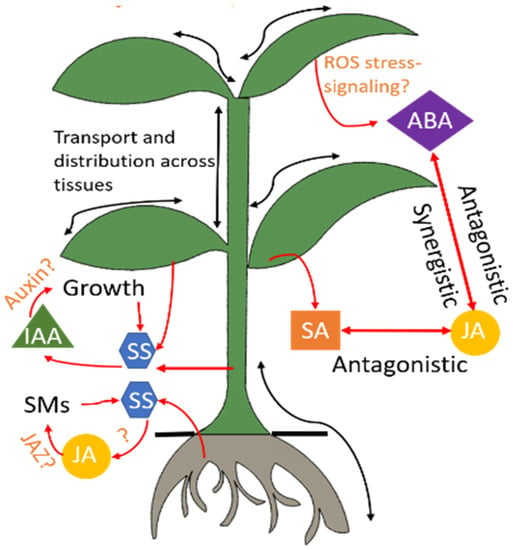
Figure 2 shows a simplified model of the phytohormone dynamics in response to a stress condition
[37]. SA, JA, ABA, and IAA are among the most important regulators of induced defense mechanisms
[33][36][38][39][40][41][42][43][33,36,38,39,40,41,42,43]. Progressive variations in their levels have been reported in response to abiotic stressors including drought, salt, and cold/heat conditions
[43][44][45][46][43,44,45,46] as well as biotic stressors including pathogen infection
[47][48][47,48]. Exogenous application of these hormones is found to mitigate oxidative stress in plants
[46][49][50][51][52][53][54][55][46,49,50,51,52,53,54,55]. Oxidative stress occurs due to a burst of reactive oxygen species, which are triggered by biotic (attack by microbes, pests, herbivores) and abiotic (drought/floods, temperature variations, soil nutrient/salinity/pH deficiencies) stresses. Root exudates are another form of signaling molecules that mediate the communication of plants with the rhizosphere. Root exudates are primarily composed of sugars, amino acids, organic acids, and vitamins, serving as a rich source of nutrients for the microbial community
[56]. These exudates serve as carbon and energy sources for microorganisms living in the rhizosphere, while also profoundly influencing the composition and diversity of the microbial community
[57]. Plants release the majority of photosynthates (the products of photosynthesis) into the rhizosphere through roots. Plants also release 100 teragrams (Tg) of methanol and 530 teragrams (Tg) of isoprene each year
[58][59][58,59]. The interconnected signaling pathways of the compounds secreted from plants are central to the plant’s ability to fine-tune the rhizosphere’s microbiome structure or the induction of defenses in response to stressors. This section describes the various signaling molecules that mediate plant-microbial interactions.
Figure 2. Hormonal regulation in the plant. Stress conditions trigger a cascade of phytohormones. Several signaling pathways of these hormones are still under research and hence indicated by the symbol “?.” Advanced sensors will advance understanding of these interactions through in-situ and real-time monitoring of these hormones. ABA: abscisic acid, IAA: indole-3-acetic acid, JA: jasmonic acid, JAZ: jasmonate, ROS: reactive oxygen species, SA: salicylic acid, SM: secondary metabolites, SS: soluble sugars.