Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Sirius Huang and Version 1 by Zhouxiao Li.
Thanks to the rapid development of computer-based systems and deep-learning-based algorithms, artificial intelligence (AI) has long been integrated into the healthcare field. AI is also particularly helpful in image recognition, surgical assistance and basic research. Due to the unique nature of dermatology, AI-aided dermatological diagnosis based on image recognition has become a modern focus and future trend.
- deep learning
- pattern recognition
- dermatology
- skin cancer
- intelligent diagnosis
- 3D imaging
1. Relevant Concept of AI in Dermatology
Knowledge representation and knowledge engineering are central to classical AI research [25,40][1][2]. Machine learning and its sub-field deep learning are foundations of the AI framework. “Machine Learning” refers to the automatic improvement of AI algorithms through experience and massive historical data (training datasets) to build models based on datasets that allow the algorithm to generate prediction and make decisions without programming [41][3]. “Deep learning” is a division of machine learning founded on artificial neural networks (ANNs) and representation learning. The ANN is a mathematical model that simulates the structure and function of biological neural networks, and an adaptive system with learning capabilities. The performance of an ANN depends on the number and structure of its neural layers and training dataset [42,43][4][5]. Deep learning is already widely used to detect and classify skin cancers and other skin lesions [44,45,46][6][7][8]. The most prominent deep learning networks can be divided into recursive neural networks (RvNNs), recurrent neural networks (RNNs), Kohonen self-organizing neural networks (KNNs), generative adversarial neural networks (GANs) and convolutional neural networks (CNNs) [47][9]. CNNs, a subtype of ANNs, are most frequently used for image processing and detection in medicine, particularly in dermatology, pathology and radiology [48][10]. Currently, the most implemented CNN architectures in the field of dermatology are GoogleNet, Inception-V3, V4, ResNet, Inception-ResNet V2 and Dense Net [47][9]. As the raw data source for training CNN architectures for applying deep learning, image sets with a large number of high-quality images are decisive for the diagnostic accuracy, sensitivity and specificity of the final trained AI algorithm [49][11]. An image set can be used to be managed for image data. The object contains a description of the image, the location of the image and the number of images in the set [50][12]. The most common image sets used to train AI CAD systems in dermatology today are ISIC archives (2016–2021), HAM10000, BCN20000 and PH2 image sets [51,52,53,54,55,56][13][14][15][16][17][18]. The concepts and components related to AI in the dermatology field are displayed systematically in Table 1.
Table 1. Essential terminologies involved in AI in dermatology.
| Terminology | Paraphrase |
|---|---|
| Artificial Intelligence (AI) | The intelligence manifested by machines made by humans, i.e., the ability of the machine to simulate natural intelligence. |
| Knowledge Representation | It is the field of AI dedicated to representing information about the world in a form that a computer system can utilize to solve complex tasks such as diagnosing a medical condition or having a dialog in a natural language. |
| Representation Learning (Feature Learning) | A set of techniques that allows a system to automatically discover the representations needed for feature detection or classification from raw data. |
| Machine Learning | The study of computer algorithms that improve automatically through experience. The algorithms use computational methods to learn from data without being explicitly programmed. |
| Deep Learning | A branch of machine learning methods based on artificial neural networks with representation learning. |
| Supervised Learning | Refers to the machine learning task of learning a function that maps an input to an output based on example input–output pairs. It infers a function from labeled training data consisting of a set of training examples. |
| Transfer Learning | Transfer learning is a machine learning model that allows a model developed from one task to be transferred for another task after fine-tuning and augmentation. |
| Artificial Neural Networks (ANNs) | ANNs, usually simply called neural networks (NNs), are computing systems vaguely inspired by the biological neural networks that constitute animal brains. An ANN is based on a collection of connected units or nodes called artificial neurons, which loosely model the neurons in a biological brain |
| Convolutional Neural Networks (CNNs) | CNNs are a class of neural networks; they are feedforward neural networks. Their artificial neurons can respond to a part of the surrounding units in the coverage area, most commonly applied to analyzing visual imagery. |
| Generative Adversarial Networks (GANs) | GANs are a method of unsupervised learning that learn by playing two neural networks against each other. |
| Pattern Recognition | The automated recognition of patterns and regularities in data. The environment and objects are collectively referred to as patterns. |
| Image Set | An object stores information about an image data set or a collection of image data sets. It contains image descriptions, locations of images and the number of images in the collection. |
2. The Implementation of AI in Dermatology
The diagnosis of skin diseases is mainly based on the characteristics of the lesions [57][19]. However, there are more than 2000 different types of dermatological diseases, and some skin lesions of different diseases show similarities, which makes antidiastole difficult [58][20]. At present, the global shortage of dermatologists is increasing with the high incidence of skin diseases. There is a serious deficit of dermatologists and uneven distribution, especially the developing countries and remote areas, which urgently require more medical facility, professional consultation and clinical assistance [59,60][21][22]. Rapid iteration in big data, image recognition technology and the widespread use of smartphones worldwide may be creating the largest transformational opportunity for skin diseases’ diagnosis and treatment in this era [61,62][23][24]. In addition to addressing the needs of underserved areas and the poor, AI now has the ability to provide rapid diagnoses, leading to more diverse and accessible treatments approaches [63][25]. An AI-aided system and algorithm will quickly turn out to be normal diagnosis and evaluation-related techniques. The morphological analysis of a lesion is the classic basis of dermatological diagnostics, and the face recognition and aesthetic analysis from AI have also matured and become more reliable [64,65][26][27]. Currently, some applications of AI in dermatology have already found their way into clinical practice. Specific implementation of AI in dermatology is visualized with a mind map (Figure 1) [53,66,67][15][28][29]. AI systems based on a deep learning algorithm use plentiful public skin lesion image datasets to distinguish between benign and malignant skin cancers. These datasets contain massive original images in diverse modalities, such as dermoscopy, clinical photographs or histopathological images [68][30]. In addition, deep learning was used to process the disagreements of human annotations for skin lesion images. An ensemble of Bayesian fully convolutional networks (FCNs) trained with ISIC archive was applied for the lesion image’s segmentation by considering two major factors in the aggregations of multiple truth annotations. The FCNs implemented a robust-to-annotation noise learning scheme to leverage multiple experts’ opinions towards improving the generalization performance using all available annotations efficiently [69][31]. Currently, the most representative and commonly used AI model is the CNN. It transmits input data through a series of interconnected nodes that resemble biological neurons. Each node is a unit of mathematical operation, a group of interconnected nodes in the network is called a layer and multiple layers build the overall framework of the network (Figure 2) [70,71][32][33]. Deep CNNs have also been applied to the automatic understanding of skin lesion images in recent years. Mirikharaji et al., proposed a new framework for training fully convolutional segmentation networks from a large number of cheap unreliable annotations, as well as a small fraction of expert clean annotations to handle both clean and noisy pixel-level annotations accordingly in the loss function. The results show that their spatially adaptive re-weighting method can significantly decrease the requirement for the careful labelling of images without sacrificing segmentation accuracy [72][34].
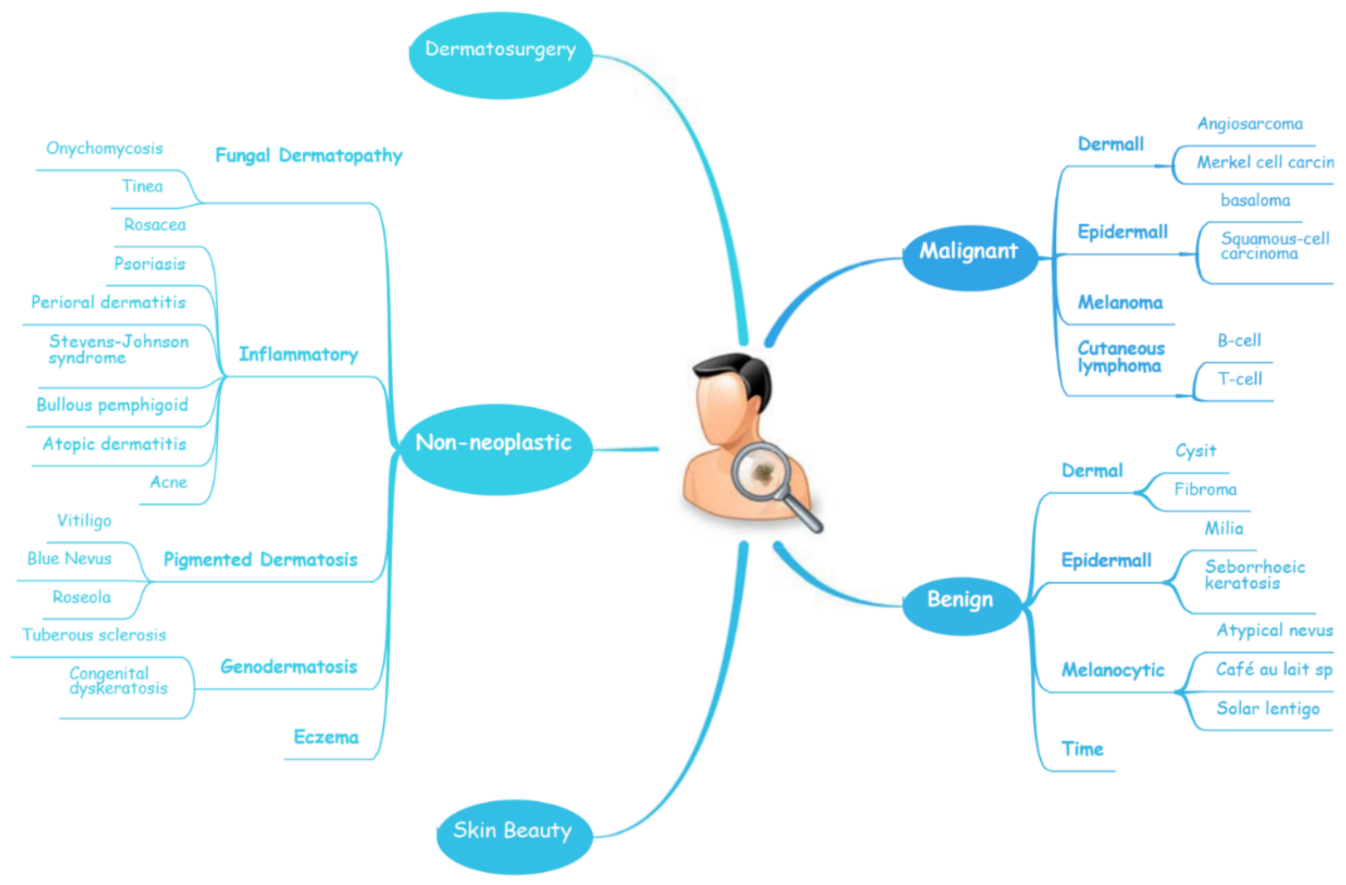
Figure 1.
A schematic illustrates the hierarchy of the implementation of AI in dermatology.
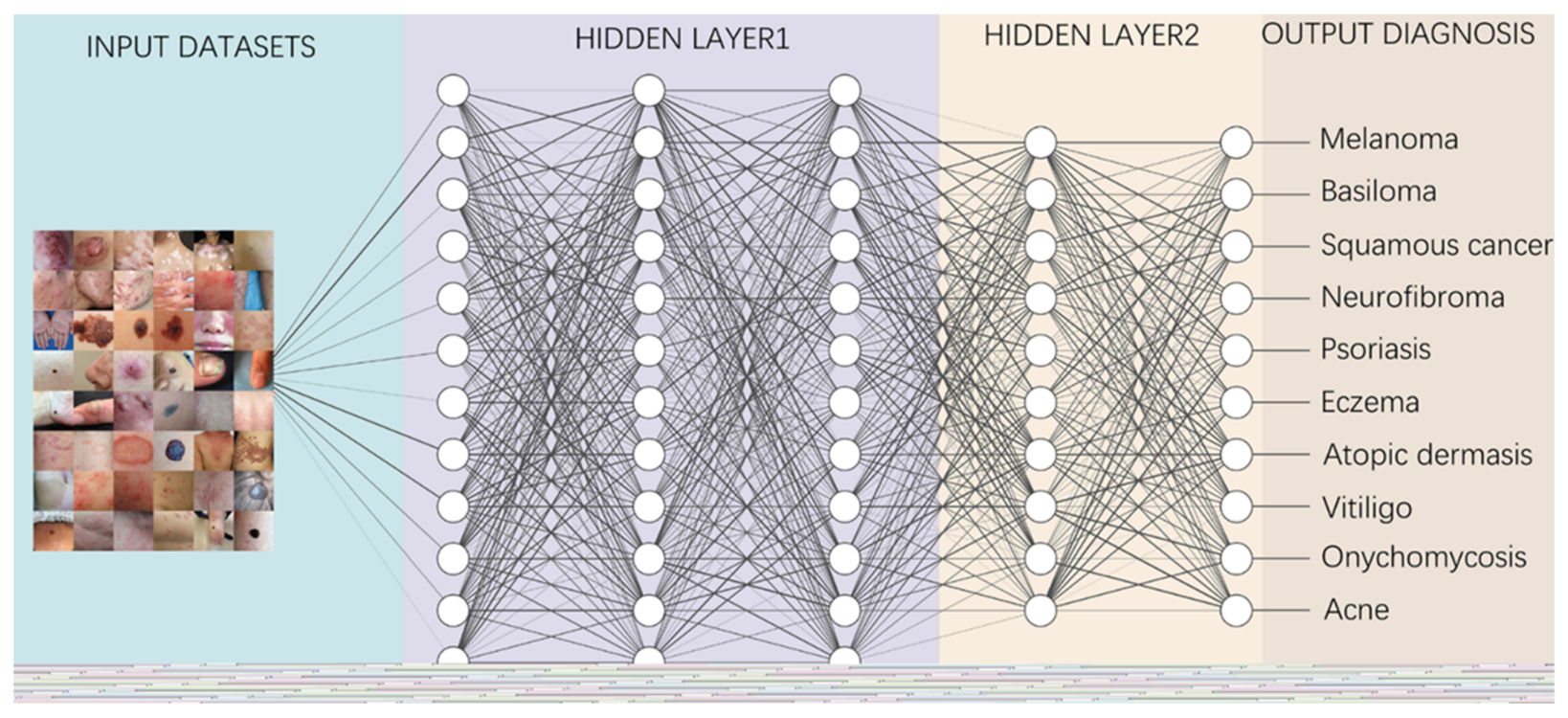
Figure 2.
A diagram depicting how to perform classification tasks in an AI neural network.
Information from the image data set is transmitted through a structure composed of multi-layer connection nodes. Each line is a weight connecting one layer to the next, with each circle representing an input, neuron or output. In convolutional neural networks, these layers contain unique convolutional layers that act as filters. The network made up of many layered filters learn increasingly high-level representations of the image.
3. AI in Aid-Diagnosis and Multi-Classification for Skin Lesions
3.1. Multi-Classification for Skin Lesions in ISIC Challenges
In recent years, the classification of multiple skin lesions has become a hotspot with the increasing popularity of using deep learning algorithms in medical image analysis. Before, metadata indicating information such as site, age, gender, etc., were not included, even though this information is collected by doctors in daily clinical practice and has an impact on their diagnostic decisions. Therefore, the algorithm or AI system that includes this information is better able to reproduce the actual diagnostic scenario, and its diagnostic performance will be more credible. The ISIC challenges consider AI systems that can identify the presence of many different pathologies and provide metadata for labelled cases, thus allowing for a more realistic comparison between AI systems and clinical scenarios. Since the International Skin Imaging Collaboration (ISIC) challenge was held in 2016, it represents the benchmark for diverse research groups working in this area. To date, their database has accumulated over 80,000 labelled training and testing images, which are openly accessible to all researchers and have been used for training algorithms to diagnose and classify various skin lesions [109][35]. In ISIC 2016–2018, subsets of the image datasets were divided into seven classes: (1) actinic keratosis and intraepithelial carcinoma, (2) basal cell carcinoma, (3) benign keratosis, (4) dermatofibroma, (5) melanocytic nevi, (6) melanoma and (7) vascular skin lesion. From 2019, the atypical nevi were added as the eighth subset. Garcia-Arroyo and Garcia-Zapirain designed a CAD system to participate in ISIC 2016, 2017 Challenge and were ranked 9th and 15th, respectively [110][36]. In 2018, Rezvantalab et al., investigated the effectiveness and capability of four pre-trained algorithms with HAM10000 (comprising a large part of the ISIC datasets) and PH2 state-of-the-art architectures (DenseNet 201, ResNet 152, Inception v3, Inception ResNet v2) in the classification of eight skin diseases. Their overall results show that all deep learning models outperform dermatologists (by at least 11%) [52][14]. Iqbal et al., proposed a deep convolutional neural network (DNN) model trained using ISIC 2017–2019 datasets that proved to be able to automatically and efficiently classify skin lesions with 0.964 AUR in ROC curve [71][33]. Similarly, Lucius’ team developed a DNN trained with HM10000 to classify seven types of skin lesions. Statistics showed that the diagnostic accuracy of dermatologists is significantly improved with the help of DNNs [111][37]. MINAGAWA et al., trained a DNN using ISIC-2017, HAM10000 and Shinshu datasets to narrow the diagnostic accuracy gap for dermatologists facing patients from different regions [112][38]. Qin et al., established a skin lesion style-based generative adversarial network (GAN) and tested it in the ISIC 2018 dataset, showing that the GAN can efficiently generate high-quality images of skin lesions, resulting in an improved performance of the classification model [73][39]. Cano et al., applied CNNs based on NASNet architecture trained with a skin image lesion from the ISIC archive for multiple skin lesion classification, which has been cross validated. Its excellent performance suggests that it can be utilized as a novel classification system for multiple classes of skin diseases [74][40]. Al-masni et al., integrated a deep learning full-resolution convolutional network and a convolutional neural network classifier for segmenting and classifying various skin lesions. The proposed integrated deep learning model was evaluated in ISIC 2016–2018 datasets and achieved an over 80% accuracy in all three for segmentation and discrimination among seven classes of skin lesions, with the highest accuracy of 89.28% in ISIC 2018 [113][41]. In 2018, Gessert et al., employed an ensemble of CNNs in the ISIC 2018 challenge and achieved second place [53][15]. Next year, they exploited a set of deep learning models trained with BCN20000 and HAM10000 datasets to solve the skin lesion classification problem, including EfficientNets, SENet and ResNeXt WSL to address the classification of skin lesions and predict unknown classes by analyzing patients’ metadata. Their approach achieved first place in the ISIC 2019 challenge [54][16].
In recent years, transfer learning technology has also been applied for classifying multiple skin lesions. Transfer learning allows a model developed from one task to be transferred for another task after fine-tuning and augmentation. It is very helpful when we don’t have enough training data sources. When lesion images are difficult to acquire, the algorithmic model can be initially performed with natural images and subsequently fine-tuned with an enhanced lesion dataset to increase the accuracy and specificity of the algorithm, thereby improving the performance on image processing tasks. Singhal et al., utilized transfer learning to train four different state-of-the-art architectures with the ISIC 2018 dataset and demonstrated their practicability for the detection of skin lesions [114][42]. Barhoumi et al., trained content-based dermatological lesion retrieval (CBDLR) systems using transfer learning, and their results showed that it outperformed a similar CBDLR systems using standard distances [75][43]. There are also some more studies that have devised AI systems or architectures trained or tested in ISIC datasets and that have gained outstanding performances [23,68,76,77,78,79,80,115,116][30][44][45][46][47][48][49][50][51].
Lately, the ISIC-2021 datasets have just been released. Except for the ISIC 18, ISIC 2019 and ISIC 2020 melanoma datasets, it also contains extra seven datasets with a total of approximately 30,000 images, such as Fitzpatric 17k, PAD-UFS-20, Derma7pt and Dermofit Image. This greatly increases the richness and diversity of the ISIC-2021 archive and correlates the patient’s skin lesion condition with the other disorders of the body, which will provide the basis for the future training of AI algorithms with a more comprehensive and higher diagnostic accuracy. Researchers are also looking forward to the publication of high-quality papers based on this archive [117][52].
3.2. Multi-Classification for Skin Lesions in Specific Dermatosis
In addition to the eight major categories of skin diseases defined in the ISIC challenge, in many specific skin diseases, a differential diagnosis for multiple subtypes is also an urgent issue to be solved. For example, in melanoma, while the common melanoma subtypes superficial spreading melanoma (SSM) and lentigo maligna melanoma (LMM) are relatively easy to diagnose, the morphological features of melanomas on other specific anatomical sites (e.g., mucosa, limb skin and nail units) are often overlooked [81][53]. On top on that, some benign nevus of melanocytic origin can also be easily confused with malignant melanoma in morphology [118][54]. Among the common pigmentation disorders, many are caused by abnormalities in melanin in the skin. Although they are similar in appearance, they are diseases with different pathological structures and treatment strategies. Diagnostic models based on AI algorithms can improve the diagnostic accuracy and specificity of these diseases so as to benefit dermatologists by reducing the time and financial cost of the diagnosis [119][55].
Melanocytic Skin Lesions
Since Binder’s team applied an ANN to discriminate between benign naevi and malignant melanoma in 1994, increasing numbers of AI algorithms are employed for the multi-classification of melanocytic skin lesions [82][56]. Moleanalyzer pro is a proven commercial CNN system for the classification of melanogenic lesions. Winkler and his team used the system, which was trained with more than 150,000 images, to investigate its diagnostic performance across different melanoma localizations and subtypes in six benign/malignant dermoscopic image sets. The CNN showed a high-level performance in most sets, except for the melanoma in mucosal and subungual sites, suggesting that the CNN may partly offset the impact of a reduced human accuracy [81][53]. In two studies by HA Haenssle et al., in 2018 and 2020, CNNs were also used in comparison with specialist dermatologists to detect melanocytic/non-melanocytic skin cancers and benign lesions. In 2018, the CNN trained with Google’s Inception v4 CNN architecture was compared with 58 physicians. The results showed that most dermatologists outperformed the CNN, but the CNN ROC curves revealed a higher specificity and doctors may benefit from assistance by a CNN’s image classification [55][17]. In 2020, Moleanalyzer pro was compared with 96 dermatologists. Even though dermatologists accomplish better results when they have richer clinical and textual case information, the overall results show that the CNN and most dermatologists perform at the same level in less artificial conditions and a wider range of diagnoses [56][18]. Sies et al., utilize the Moleanalyzer pro and Moleanalyzer daynamole systems for the classification of melanoma, melanocytic nervus and other dermatomas. The results showed that the two market-approved CAD systems offer a significantly superior diagnostic performance compared to conventional image analyzers without AI algorithms (CIA) [83][57].
Benign Pigmented Skin Lesions
Based on a wealth of experience and successful clinical practice, scholars have gradually tried to apply AI to differentiate a variety of pigmented skin diseases with promising results. Lin’s team pioneered the use of deep learning to diagnose common benign pigmented disorders. They developed two CNN models (DenseNet-96 and ResNet-152) to identify six facial pigmented dermatoses (the nevus of Ota, acquired nevus of Ota, chloasma, freckles, seborrheic keratosis and cafe-au-lait spots).Then, they introduced ResNet.99 to build a fusion network, and evaluated the performance of the two CNN with fusion networks separately. The results showed that the fusion network performance was the best and could reach a level comparable to that of dermatologists [84][58]. In 2019, Tschandl et al., conducted the world largest comparison study between the machine-learning algorithm and 511 dermatologists for the diagnosis accuracy of pigmented skin lesion classification. The algorithm was, on average, 2.01% more correct in its diagnosis compared to all human readers. The result disclosed that machine-learning classifiers outperform dermatologist in the diagnosis of skin pigmented lesions and should be more widely used in clinical practice [120][59]. In the latest study, Lyakhov et al., established a multimodal neural network for the hair removal preliminary process and differentiation of the 10 most common pigmented lesions (7 benign and 3 malignant). They found that fusing metadata from various sources could provide additional information, thereby improving the efficiency of the neural network analysis and classification system, as well as the accuracy of the diagnosis. Experimental results showed that the fusion of metadata led to an increase in recognition accuracy of 4.93–6.28%, with a maximum diagnosis rate of 83.56%. The study demonstrated that the fusion of patient statistics and visual data makes it possible to find extra connections between dermatoscopic images and medical diagnoses, significantly improving the accuracy of neural network classification [85][60].
Inflammatory Dermatoses
Inflammatory dermatoses are a group of diseases caused by the destruction of skin tissue as a result of immune system disorders, including eczema, atopic dermatitis, psoriasis, chronic urticarial and pemphigus. Newly recorded histological findings and neoteric applications of immunohistochemistry have also refined the diagnosis of inflammatory skin diseases [121][61]. AI CAD systems are able to optimize the workflow of highly routinely diagnosed inflammatory dermatoses. A multi-model, multi-level system using an ANN architecture was designed for eczema detection. This system is conceived as an architecture with different models matching input features, and the output of these models are integrated through a multi-level decision layer to calculate the probability of eczema, resulting in a system with a higher confidence level than a single-level system [86][62]. From 2017 onwards, neural networks have been shown to be useful for diagnosing acne vulgaris [90][63]. The latest publications on the use of computer-aided systems in acne vulgaris are based on a wealth of data from cell phone photographs of affected patients, which enable the development of AI-based algorithms to determine the severity of facial acne and to identify different types of acne lesions or post-inflammatory hyperpigmentation [91][64]. Scientists in South Korea trained various image analysis algorithms to recognize images of fungal nails. For this purpose, they used datasets of almost 50,000 nail images and 4 validation datasets of a total of 1358 images. A comparison of the respective diagnostic accuracy (measured in this study by the Youden index) of differently trained assessors and the AI algorithm showed the highest diagnostic accuracy in the computer-based image analysis and was significant superior to dermatologists (p = 0.01) [87][65].
4. AI in Aid-Diagnosis and Binary-Classification for Specific Dermatosis
4.1. Skin Cancer
The incidence of skin cancer has been increasing yearly [58,122][20][66]. Although its mortality rate is relatively low [123][67], it remains a heavy economic burden on health services and can cause severe mental problems, especially as most skin cancers occur in highly visible areas of the body [124][68]. Due to the low screening awareness, a lack of specific lesion features in early skin cancer and insufficient adequate clinical expertise and services, most patients were only diagnosed at an advanced stage, thus leading to a poor prognosis [124[68][69],125], so there is an urgent need for AI systems to help clinicians in this field.
Melanoma
Melanoma is the deadliest type of skin cancer. The early screening and early diagnosis of melanoma is essential to improve patient survival [126][70]. Currently, dermatologists diagnose melanoma mainly by applying the ABCD principle based on the morphological characteristics of melanoma lesions [127][71]. However, even for experienced dermatologists, this manual examination is non-trivial, time consuming and can be easily confused with other benign skin lesions [128][72]. Thus, most AI-driven skin cancer research has focused on the classification of melanocytic lesions to aid melanoma screening. In 2004, Blum et al., pioneered the use of computer algorithms for the diagnosis of cutaneous melanoma and proved that a diagnostic algorithm for the digital image analysis of melanocytic diseases could achieve a similar accuracy to expert dermatoscopy [88][73]. In 2017, Esteva et al., trained a GoogleNet-Inception-v3-based CNN with the training dataset, including 129,450 clinical images of 2032 different diseases from 18 sites. The performance of the CNN was compared with 21 dermatologists in two critical binary classifications (the most common cancer and the deadliest skin cancer) of biopsy-confirmed clinical images. The CNN’s performance on both tasks was competent, and comparable to that of dermatologists, demonstrating its ability to classify skin cancer [66][28]. The ISIC Melanoma Project has also created a publicly accessible archive of images of skin lesions for education and research. Marchetti et al., summarized the results of a melanoma classification for ISIC challenge in 2016, which involved 25 competing teams. They compared the algorithm’s diagnosis with those of eight experienced dermatologists. The outcomes showed that automated algorithms significantly outperformed the dermatologists in diagnosing melanoma [89][74]. Subsequently, they made a comparison of the computer algorithms’ performance of 32 teams in the ISIC 2017 challenge with 17 human readers. The results also demonstrated that deep neural networks could classify skin images of melanoma and its benign simulants with a high precision and have the potential to boost the performance of human readers [22][75]. Filho and Tangs’ team have utilized the ISIC 2016, 2017 challenge and PH2 datasets to develop the algorithm for the classification and segmentation of the melanoma area automatically. Their test outcomes indicated that these algorithms could dramatically improve the doctors’ efficiency in diagnosing melanoma [51,129][13][76]. In MacLellan’s study, three AI-aid diagnosis systems were compared with dermatologists using 209 lesions in 184 patients. The statistics showed that the Moleanalyzer pro had a relative high sensitivity and the highest specificity (88.1%, 78.8%), whereas local dermatologists had the highest sensitivity but a low specificity (96.6%, 32.2%) [130][77]. Consistently, Moleanalyzer pro also showed its reliability in the differentiation of combined naevi and melanomas [131][78]. It is also possible for dermatologists to build a whole-body map using a 3D imaging AI system; its application is of particular relevance in the context of skin cancer diagnostics. The 360° scanner uses whole-body images to create a “map” of pigmented skin lesions. Using a dermatoscope, atypical and altered nevi can also be examined microscopically and stored digitally. With the help of intelligent software, emerging lesions or lesions that change over time are automatically marked during follow-up checks—an important feature for recognizing a malignancy and initiating therapeutic measures [132][79]. In addition, in the long term, high-risk melanoma populations will benefit from a clinical management approach that combines an AI-based 3D total-body photography monitor with sequential digital dermoscopy imaging and teledermatologist evaluation [133,134][80][81].
Non-Melanoma Skin Cancer
AI is also widely used to differentiate between malignant and benign skin lesions, along with the detection of non-melanoma skin cancer (NMSC). Rofman et al., proposed a multi-parameter ANN system based on personal health management data that can be used to forecast and analyze the risk of NMSC. The system was trained and validated by 2056 NMSC and 460,574 non-cancer cases from the 1997–2015 NHIS adult survey data, and was then further tested by 28058 individuals from the 2016 NHIS survey data. The ANN system is available for the risk assessment of non-melanoma skin cancer with a high sensitivity (88.5%). It can classify patients into high, medium and low cancer risk categories to provide clinical decision support and personalized cancer risk management. The study’s model is therefore a prediction, where clinicians can obtain information and the patient risk status to detect and prevent non-melanoma skin cancer at an early stage [94][82]. Alzubaidi et al., propose a novel approach to overcome the lack of enough input-labeled raw skin lesion images by retraining a deep learning model based on large unlabeled medical images on a small number of labeled medical images through transfer learning. The model has an F1-score value of 98.53% in distinguishing skin cancer from normal skin [95][83].
Neurofibroma
Neurofibromatosis (NF) is a group of three conditions in which tumors grow in the nervous system, and are NF1, NF2 and schwannomatosis [135][84]. NF1 is the most common neurofibroma and cancer susceptibility disease. Most patients with NF1 have a normal life expectancy, but 10% of them develop malignant peripheral nerve sheath tumors (MPNST), which is a major cause of morbidity [136][85]. Therefore, the timely differentiation of benign and malignant lesions has direct significance for improving the survival rate of patients. Wei et al., successfully established a Keras-based machine-learning model that can discriminate between NF1-related benign and malignant craniofacial lesions with a very high accuracy (96.99 and 100%) in validation cohorts 1 and 2 and a 51.27% accuracy in various other body regions [137][86]. Plexiform neurofibroma (PN) is a prototypical and most common NF1 tumor. Ho et al., created a DNN algorithm to conduct a semi-automated volume segmentation of PNs based on multiple b-value diffusion-weighted MRI. They evaluated the accuracy of semi-automated tumor volume maps constructed by a DNN compared to manual segmentation and revealed that the volumes generated by the DNN from multiple diffusion data on PNs have a good correlation with manual volumes, and that there is a significance between PN and normal tissue [97][87]. Interestingly, Bashat and his colleagues also demonstrated that a quantitative image representation method based on machine learning may assist in the classification between benign PNs and MPNST in NF1 [102][88]. In a similar initiative, Duarte et al., used grey matter density maps obtained from magnetic resonance (MR) brain structure scans to create a multivariate pattern analysis algorithm to differentiate between NF1 patients and healthy controls. A total of 83% of participants were correctly classified, with 82% sensitivity and 84% specificity, demonstrating that multivariate techniques are a useful and powerful tool [103][89].
4.2. Application of AI for Inflammatory Dermatosis
Psoriasis
The prevalence of psoriasis is 0% to 2.1% in children and 0.91% to 8.5% in adults [138][90]. The psoriasis area and severity index (PASI), body surface area (BSA) and physician global assessment (PGA) are the three most commonly used indicators to evaluate psoriasis severity [139,140][91][92]. However, both PASI and BSA have been repeatedly questioned for their objectivity and reliability [141][93]. It would therefore be of great help to use AI algorithms to make a standardized and objective assessment. Nowadays, machine-learning-based algorithms are available to determine BSA scores. Although this algorithm had slight limitations in detecting flaking as diseased skin, it has reached an expert level in BSA assessment [104][94]. At present, there are already computer-assisted programs for PASI evaluation, which, however, still require human assistance and function by recognizing predefined threshold values for certain characteristics [98][95]. Another study by Fink’s team is also based on image analysis with the FotoFinderTM. The accuracy and reproducibility of PASI has been impressively improved with the help of semi-automatic computer-aided algorithms [99][96]. These technological advances in BSA and PASI measurements are expected to greatly reduce the workload of doctors while ensuring a high degree of repeatability and standardization. In addition to the three above indicators, Anabik Pal et al., used erythema, scaling and induration to build a DNN to determine the severity of psoriatic plagues. The algorithm is given a psoriasis image and then makes a prediction about the severity of the three parameters. This task is seen as a new multi-task learning (MTL) problem formed by three interdependent subtasks in addition to three different single task learning (STL) problems, so the DNN is trained accordingly. The training dataset consists of 707 photographs and the training results show that the deep CNN-based MTL approach performs well when grading the disease parameters alone, but slightly less well when all three parameters are correctly graded at the same time [142][97].
AI can also assist in evaluating and diagnosing psoriasis. Munro’s microabscesses (MM) is a sign of psoriasis. Anabik Pak et al., presented a computational framework (MICaps) to detect neutrophils in the epidermal stratum corneum of the skin from biopsy images (a component of MM detection in skin biopsies). Using MICaps, the diagnosis performance was increased by 3.27% and model parameters were reduced by 50% [143][98]. A CNN algorithm that differentiated among nine diagnoses based on photos made fewer misdiagnoses and had a lower omission diagnostic rate of psoriasis compared to 25 dermatologists [92][99]. In addition, Emma et al., used machine learning to find out which psoriasis patient characteristics are associated with long-term responses to biologics [144][100]. Thanks to AI, an amelioration in diagnosis and treatment can be inferred in psoriasis patients.
Eczema
The challenge in the computer-aided image diagnosis of eczematous diseases is to correctly differentiate not only between disease and health, but also between different forms of eczema. The eczema stage and affected area are the most essential factors in effectively assessing the dynamics of the disease. It is not trivial to accurately identify the eczema area and other inflammatory dermatoses on the basis of photographic documentation. The macroscopic forms of eczema are diverse, with different stages and varying degrees of distribution and severity [145][101]. The prerequisite for training algorithms for the AI-supported image analysis of all of these various assessment parameters is therefore a correspondingly large initial quantity of image files that have been optimized and adjusted in terms of the recording technology. Forms of eczema with disseminated eruption, such as the corresponding manifestation patterns of atopic dermatitis, would also be linked to the availability of automated digital, serial whole-body photography for an efficient and time-saving AI-supported calculation of an area score. Han et al., trained a deep neural-network-based algorithm. The algorithm is able to differentiate between eczema and other infectious skin diseases and to classify very rare skin lesions, which has direct clinical significance, and to serve as augmented intelligence to empower medical professionals in diagnostic dermatology. They even showed that treatment recommendations (e.g., topical steroids versus antiseptics) could also be learned by differentiating between inflammatory and infectious causes. It remains to be seen and questioned, however, whether an AI-aided severity assessment and a clinically practicable area score can be derived from this as a prerequisite for a valid follow-up in the case of eczema [93][102]. Schnuerle et al., designed a support-vector-machine-based image processing method for hand eczema segmentation with industry swiss4ward for operational use at the University Hospital Zurich. This system uses the F1-score as the primary measurement and is superior to a few advanced methods that were tested on their gold standard dataset likewise [100][103]. Presumably, a combination of such an AI-aided image analysis and molecular diagnostics can optimize the future differential diagnostic classification of eczema diseases, as recently predicted for various clinical manifestations of hand dermatitis [146][104].
Atopic Dermatitis
Atopic dermatitis (AD) is the most common chronic inflammatory disease, with a prevalence of 10% to 20% in developed countries [147][105]. It usually starts in childhood and recurs multiple times in adulthood, greatly affecting patients’ quality of life [148][106]. In 2017, Gustofson’s team identified patients with AD via a machine-learning-based phenotype algorithm. The algorithm combined code information with the collection of electronic health records to achieve a high positive predictive value and sensitivity. These results demonstrate the utility of natural language processing and machine learning in EHR-based phenotyping [105][107]. An ANN algorithm was developed to assess the influence of air contaminants and weather variation on AD patients; their results proved that the severity of AD symptoms was positively correlated with outdoor temperatures, RH, precipitation, NO2, O3 and PM10 [149][108]. In the latest study, a fully automatic approach based on CNN was proposed to analysis multiphoton tomography (MPT) data. The proposed algorithm correctly diagnosed AD in 97.0 ± 0.2% of all images presenting living cells, with a sensitivity of 0.966 ± 0.003 and specificity of 0.977 ± 0.003, indicating that MPT imaging can be combined with AI to successfully diagnose AD [96][109].
Acne
The assessment of AI has been very effective. Melina et al., showed an excellent correlation between the automatic and manual evaluation of the investigator’s global assessment with r = 0.958 [150][110]. In the case of acne vulgaris in particular, such a procedure could prevent far-reaching consequences with permanent skin damage in the form of scars.
Vitiligo
The depigmented macules of vitiligo are usually in high contrast to unaffected skin. Vitiligo is more easily recognized by AI systems than features of eczema or psoriasis lesions with poorly defined borders. Computer-based algorithms used for the detection of vitiligo with an F1 score of 0.8462 demonstrated an impressive superiority to pustular psoriasis [151][111]. Luo designed a vitiligo AI diagnosis system employing cycle-consistent adversarial networks (cycle GANs) to generate images in Wood’s lamp and improved the image resolution via an attention-aware dense net with residual deconvolution (ADRD). The system achieved a 9.32% improvement in classification performance accuracy compared to direct classification of the original images using Resnet50 [106][112]. Makena’s team built a CNN that performs vitiligo skin lesion segmentation quickly and robustly. The network was trained on 308 images with various lesion sizes, intricacies and anatomical locations. The modified network outperformed the state-of-the-art U-Net with a much higher Jaccard index score (73.6% versus 36.7%) and shorter segmentation time than the previously proposed semi-autonomous watershed approach [107][113]. These novel systems have proved promising for clinical applications by greatly saving the testing time and improving the diagnostic accuracy.
Fungal Dermatosis
Gao et al., invented an automated microscope for fungal detection in dermatology based on deep learning. The system is as proficient as a dermatologist in detecting skin and nail specimens, with sensitivities of 99.5% and 95.2% and specificities of 91.4% and 100%, respectively [101][114].
5. Application of AI for Aesthetic Dermatology
AI combined with new optical technologies is also increasingly being applied in aesthetics dermatology. Examples include face recognition, automatic beautification in smartphones and related software. So-called smart mirror analyzers are now available on the Internet, which are AI-assisted technologies with image recognition systems that analyze the skin based on its appearance and current external environment and recommend skin care products accordingly [152][115]. The program ArcSoft Protrait can automatically identify the wrinkles, moles, acne and cicatrice and intelligently soften, moisturize and smooth the skin while retaining a maximum skin texture and detail, greatly simplifying the cumbersome and time-consuming portrait process [61,64,153][23][26][116]. AI also plays an essential role in facial aesthetics assessing. For this purpose, ANNs are trained using face image material that people judge independently according to aesthetic criteria based on various criteria. The ANN learns from photos and their respective attractiveness ratings to make human-like judgments about the aesthetics of the face [65][27]. New applications objectively evaluate each photo on the basis of over 80 facial coordinates and nearly 7000 associated distances and angles [108][117].
6. Applications of AI for Skin Surgery
Radical resection and amputation are the best means of preventing recurrence and fatal metastasis for malignant dermatoma [154][118]. A skin or flap graft via microsurgery and the application of prosthesis play a crucial role in improving the quality of life of patients after resection [155,156][119][120]. Adequate microvascular anastomosis is the key to a successful microvascular-free tissue transfer. As a basic requirement in this regard, the surgeon must have excellent microsurgery skills. Thanks to the support of a series of auxiliary equipment such as microscopes, magnifications of up to 10 to 15 times are possible and allow for the anastomosis of small vessels. Nevertheless, due to physiological tremor, only vessels of up to approximately 0.5–1 mm in size can be safely anastomosed, especially in lymphatic surgery or perforator-based flaps, where the vascular caliber may even be smaller, which is why surgeons reach their limits here [157][121]. In this background, the expansion of surgical microscopes to include robotics and AI capabilities represents a promising and innovative approach for surpassing the capabilities of the human hand. The aim is to use robots equipped with AI to eliminate human tremor and to enable motion scaling for an increased precision and dexterity in the smallest of spaces [158][122]. By downscaling human movements, finer vessels can be attached. In the future, advances could be achieved in the field of ultra-microsurgery and anastomoses in the range of 0.1–0.8 mm on the smallest vessels or nerve fascicles. In the long term, intelligent robotics could also automate technically demanding tasks, such as microsurgical anastomosis performed by robots, or the implementation of a real-time feedback system for the surgeon.
Prosthetics have also evolved with the implementation of AI. After amputation injuries, prostheses can now restore not only the shape but also essential functions of the amputated extremity; in this way, they make a significant contribution to the reintegration of the patient into society. The mental control of the extremity remains in the brain even after amputation. When movement patterns are imagined, despite the lack of end organs to perform them, neurons will still transmit corresponding nerve signals [159][123]. Prostheses can now receive the electrical potential via up to eight electrodes and assign them to the respective functions via pattern recognition and innovative technological methods equipped with AI, and can ensure that patients better use the prosthesis in their daily lives [160,161][124][125]. This enables the patient to directly control different grip shapes and movements, which means that gripping movements can be realized much faster and more naturally in terms of movement behavior.
The application of AI-based surgical robots in skin surgery is now also becoming widespread. Compared to traditional open surgery, robotic-assisted surgery offers 3D vision systems and flexible operating instruments, with potentially fewer postoperative complications as a result. In 2010, Sohn et al., first applied this technique to treat two pelvic metastatic melanoma patients [162][126]. In 2017, Kim successfully treated one case of vaginal malignant melanoma using robotic-assisted anterior pelvic organ resection with ileoccystostomy [15][127]. One year later, Hyde successfully treated four cases of malignant melanoma using robotic-assisted inguinal lymph node dissection [163][128]. Miura et al., found that robotic assistance provided a safe, effective and minimally invasive method of removing a pelvic lymph from patients with peritoneal metastases melanoma, with shorter hospital stays compared to normal open surgery [164][129]. Medical robots are also involved in the field of hair transplantation. In 2011, the ARTAS system was officially approved by the US-FDA for male hair transplantation, providing clear and detailed characteristics of the donor area by capturing microscopically magnified images and computer-aided parameters to facilitate the acquisition of complete follicular units from the donor area [16][130]. The system reduces labor consumption and eliminates human fatigue and potential errors, and the procedure time is significantly reduced [165][131].
References
- Russell, S.; Norvig, P. Artificial Intelligence: A Modern Approach, 4th ed.; Prentice Hall: Hoboken, NJ, USA, 2020.
- Russell, S.J.; Norvig, P. Artificial Intelligence. In The ACM Computing Classification System 1998; Association for Computing Machinery Inc.: New York, NY, USA, 2007; pp. 320–363.
- Mitchell, T. Machine Learning, 1st ed.; McGraw-Hill Education Ltd.: New York, NY, USA, 1997.
- Yegnanarayana, B. Artificial Neural Networks, 1st ed.; Prentice-Hall of India Pvt. Ltd.: Chennai, India, 2004.
- Erickson, B.J.; Korfiatis, P.; Kline, T.L.; Akkus, Z.; Philbrick, K.; Weston, A.D. Deep Learning in Radiology: Does One Size Fit All? J. Am. Coll. Radiol. 2018, 15, 521–526.
- Manne, R.; Kantheti, S.; Kantheti, S. Classification of Skin Cancer Using Deep Learning, Convolutional Neural Networks—Opportunities and Vulnerabilities—A Systematic Review. Int. J. Mod. Trends Sci. Technol. 2020, 6, 101–108.
- Dildar, M.; Akram, S.; Irfan, M.; Khan, H.U.; Ramzan, M.; Mahmood, A.R.; Alsaiari, S.A.; Saeed, A.H.M.; Alraddadi, M.O.; Mahnashi, M.H. Skin Cancer Detection: A Review Using Deep Learning Techniques. Int. J. Environ. Res. Public Health 2021, 18, 5479.
- Fink, C.; Blum, A.; Buhl, T.; Mitteldorf, C.; Hofmann-Wellenhof, R.; Deinlein, T.; Stolz, W.; Trennheuser, L.; Cussigh, C.; Deltgen, D.; et al. Diagnostic Performance of a Deep Learning Convolutional Neural Network in the Differentiation of Combined Naevi and Melanomas. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 1355–1361.
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of Deep Learning: Concepts, CNN Architectures, Challenges, Applications, Future Directions; Springer International Publishing: Cham, Switzerland, 2021; Volume 8, ISBN 4053702100444.
- Ghosh, A.; Sufian, A.; Sultana, F.; Chakrabarti, A.; De, D. Fundamental Concepts of Convolutional Neural Network. In Recent Trends and Advances in Artificial Intelligence and Internet of Things; Springer: Berlin/Heidelberg, Germany, 2020; pp. 519–567.
- Daneshjou, R.; Vodrahalli, K.; Novoa, R.A.; Jenkins, M.; Liang, W.; Rotemberg, V.; Ko, J.; Swetter, S.M.; Bailey, E.E.; Gevaert, O. Disparities in Dermatology AI Performance on a Diverse, Curated Clinical Image Set. arXiv 2022, arXiv:2203.08807.
- Zhao, Z.-Q.; Xu, S.-T.; Liu, D.; Tian, W.-D.; Jiang, Z.-D. A Review of Image Set Classification. Neurocomputing 2019, 335, 251–260.
- Rebouças Filho, P.P.; Peixoto, S.A.; Medeiros da Nóbrega, R.V.; Hemanth, D.J.; Medeiros, A.G.; Sangaiah, A.K.; de Albuquerque, V.H.C. Automatic Histologically-Closer Classification of Skin Lesions. Comput. Med. Imaging Graph. 2018, 68, 40–54.
- Rezvantalab, A.; Safigholi, H.; Karimijeshni, S. Dermatologist Level Dermoscopy Skin Cancer Classification Using Different Deep Learning Convolutional Neural Networks Algorithms. arXiv 2018, arXiv:1810.10348.
- Gessert, N.; Sentker, T.; Madesta, F.; Schmitz, R.; Kniep, H.; Baltruschat, I.; Werner, R.; Schlaefer, A. Skin Lesion Diagnosis Using Ensembles, Unscaled Multi-Crop Evaluation and Loss Weighting. arXiv 2018, arXiv:1808.01694.
- Gessert, N.; Nielsen, M.; Shaikh, M.; Werner, R.; Schlaefer, A. Skin Lesion Classification Using Ensembles of Multi-Resolution EfficientNets with Meta Data. MethodsX 2020, 7, 100864.
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Ben Hadj Hassen, A.; Thomas, L.; Enk, A.; et al. Man against Machine: Diagnostic Performance of a Deep Learning Convolutional Neural Network for Dermoscopic Melanoma Recognition in Comparison to 58 Dermatologists. Ann. Oncol. 2018, 29, 1836–1842.
- Haenssle, H.A.; Fink, C.; Toberer, F.; Winkler, J.; Stolz, W.; Deinlein, T.; Hofmann-Wellenhof, R.; Lallas, A.; Emmert, S.; Buhl, T.; et al. Man against Machine Reloaded: Performance of a Market-Approved Convolutional Neural Network in Classifying a Broad Spectrum of Skin Lesions in Comparison with 96 Dermatologists Working under Less Artificial Conditions. Ann. Oncol. 2020, 31, 137–143.
- Berk-Krauss, J.; Polsky, D.; Stein, J.A. Mole Mapping for Management of Pigmented Skin Lesions. Dermatol. Clin. 2017, 35, 439–445.
- Demers, A.A.; Nugent, Z.; Mihalcioiu, C.; Wiseman, M.C.; Kliewer, E.V. Trends of Nonmelanoma Skin Cancer from 1960 through 2000 in a Canadian Population. J. Am. Acad. Dermatol. 2005, 53, 320–328.
- Weinberg, J.; Kaddu, S.; Gabler, G.; Kovarik, C. The African Teledermatology Project: Providing Access to Dermatologic Care and Education in Sub-Saharan Africa. Pan Afr. Med. J. 2009, 3, 16.
- Gaffney, R.; Rao, B. Global Teledermatology. Glob. Dermatol. 2015, 2, 209–214.
- Kaliyadan, F.; Ashique, K.T. Use of Mobile Applications in Dermatology. Indian J. Dermatol. 2020, 65, 371–376.
- Freeman, K.; Dinnes, J.; Chuchu, N.; Takwoingi, Y.; Bayliss, S.E.; Matin, R.N.; Jain, A.; Walter, F.M.; Williams, H.C.; Deeks, J.J. Algorithm Based Smartphone Apps to Assess Risk of Skin Cancer in Adults: Systematic Review of Diagnostic Accuracy Studies. BMJ 2020, 368, m127.
- Veronese, F.; Branciforti, F.; Zavattaro, E.; Tarantino, V.; Romano, V.; Meiburger, K.M.; Salvi, M.; Seoni, S.; Savoia, P. The Role in Teledermoscopy of an Inexpensive and Easy-to-Use Smartphone Device for the Classification of Three Types of Skin Lesions Using Convolutional Neural Networks. Diagnostics 2021, 11, 451.
- Eisentha, Y.; Dror, G.; Ruppin, E. Facial Attractiveness: Beauty and the Machine. Neural Comput. 2006, 18, 119–142.
- Kagian, A.; Dror, G.; Leyvand, T.; Meilijson, I.; Cohen-Or, D.; Ruppin, E. A Machine Learning Predictor of Facial Attractiveness Revealing Human-like Psychophysical Biases. Vis. Res. 2008, 48, 235–243.
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-Level Classification of Skin Cancer with Deep Neural Networks. Nature 2017, 542, 115–118.
- Mahbod, A.; Tschandl, P.; Langs, G.; Ecker, R.; Ellinger, I. The Effects of Skin Lesion Segmentation on the Performance of Dermatoscopic Image Classification. Comput. Methods Programs Biomed. 2020, 197, 105725.
- Mahbod, A.; Schaefer, G.; Wang, C.; Dorffner, G.; Ecker, R.; Ellinger, I. Transfer Learning Using a Multi-Scale and Multi-Network Ensemble for Skin Lesion Classification. Comput. Methods Programs Biomed. 2020, 193, 105475.
- Mirikharaji, Z.; Abhishek, K.; Izadi, S.; Hamarneh, G. D-LEMA: Deep Learning Ensembles from Multiple Annotations-Application to Skin Lesion Segmentation. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Nashville, TN, USA, 19–25 June 2021; pp. 1837–1846.
- Yang, C.-H.; Ren, J.-H.; Huang, H.-C.; Chuang, L.-Y.; Chang, P.-Y. Deep Hybrid Convolutional Neural Network for Segmentation of Melanoma Skin Lesion. Comput. Intell. Neurosci. 2021, 2021, 9409508.
- Iqbal, I.; Younus, M.; Walayat, K.; Kakar, M.U.; Ma, J. Automated Multi-Class Classification of Skin Lesions through Deep Convolutional Neural Network with Dermoscopic Images. Comput. Med. Imaging Graph. 2021, 88, 101843.
- Mirikharaji, Z.; Yan, Y.; Hamarneh, H. Learning to Segment Skin Lesions from Noisy Annotations; Springer International Publishing: Cham, Switzerland, 2019; Volume 4, ISBN 9783030333911.
- Cassidy, B.; Kendrick, C.; Brodzicki, A.; Jaworek-Korjakowska, J.; Yap, M.H. Analysis of the ISIC Image Datasets: Usage, Benchmarks and Recommendations. Med. Image Anal. 2022, 75, 102305.
- Garcia-Arroyo, J.L.; Garcia-Zapirain, B. Segmentation of Skin Lesions in Dermoscopy Images Using Fuzzy Classification of Pixels and Histogram Thresholding. Comput. Methods Programs Biomed. 2019, 168, 11–19.
- Lucius, M.; De All, J.; De All, J.A.; Belvisi, M.; Radizza, L.; Lanfranconi, M.; Lorenzatti, V.; Galmarini, C.M. Deep Neural Frameworks Improve the Accuracy of General Practitioners in the Classification of Pigmented Skin Lesions. Diagnostics 2020, 10, 969.
- Minagawa, A.; Koga, H.; Sano, T.; Matsunaga, K.; Teshima, Y.; Hamada, A.; Houjou, Y.; Okuyama, R. Dermoscopic Diagnostic Performance of Japanese Dermatologists for Skin Tumors Differs by Patient Origin: A Deep Learning Convolutional Neural Network Closes the Gap. J. Dermatol. 2021, 48, 232–236.
- Qin, Z.; Liu, Z.; Zhu, P.; Xue, Y. A GAN-Based Image Synthesis Method for Skin Lesion Classification. Comput. Methods Programs Biomed. 2020, 195, 105568.
- Cano, E.; Mendoza-Avilés, J.; Areiza, M.; Guerra, N.; Mendoza-Valdés, J.L.; Rovetto, C.A. Multi Skin Lesions Classification Using Fine-Tuning and Data-Augmentation Applying Nasnet. PeerJ Comput. Sci. 2021, 7, e371.
- Al-masni, M.A.; Kim, D.-H.H.; Kim, T.-S.S. Multiple Skin Lesions Diagnostics via Integrated Deep Convolutional Networks for Segmentation and Classification. Comput. Methods Programs Biomed. 2020, 190, 105351.
- Singhal, A.; Shukla, R.; Kankar, P.K.; Dubey, S.; Singh, S.; Pachori, R.B. Comparing the Capabilities of Transfer Learning Models to Detect Skin Lesion in Humans. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 2020, 234, 1083–1093.
- Barhoumi, W.; Khelifa, A. Skin Lesion Image Retrieval Using Transfer Learning-Based Approach for Query-Driven Distance Recommendation. Comput. Biol. Med. 2021, 137, 104825.
- Kassem, M.A.; Hosny, K.M.; Fouad, M.M. Skin Lesions Classification into Eight Classes for ISIC 2019 Using Deep Convolutional Neural Network and Transfer Learning. IEEE Access 2020, 8, 114822–114832.
- Ratul, M.A.R.; Mozaffari, M.H.; Lee, W.-S.; Parimbelli, E. Skin Lesions Classification Using Deep Learning Based on Dilated Convolution. bioRxiv 2020.
- Rashid, H.; Tanveer, M.A.; Aqeel Khan, H. Skin Lesion Classification Using GAN Based Data Augmentation. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 916–919.
- Maron, R.C.; Weichenthal, M.; Utikal, J.S.; Hekler, A.; Berking, C.; Hauschild, A.; Enk, A.H.; Haferkamp, S.; Klode, J.; Schadendorf, D.; et al. Systematic Outperformance of 112 Dermatologists in Multiclass Skin Cancer Image Classification by Convolutional Neural Networks. Eur. J. Cancer 2019, 119, 57–65.
- Sun, Q.; Huang, C.; Chen, M.; Xu, H.; Yang, Y. Skin Lesion Classification Using Additional Patient Information. BioMed Res. Int. 2021, 2021, 6673852.
- Jain, S.; Singhania, U.; Tripathy, B.; Nasr, E.A.; Aboudaif, M.K.; Kamrani, A.K. Deep Learning-Based Transfer Learning for Classification of Skin Cancer. Sensors 2021, 21, 8142.
- Le, D.N.T.; Le, H.X.; Ngo, L.T.; Ngo, H.T. Transfer Learning with Class-Weighted and Focal Loss Function for Automatic Skin Cancer Classification. arXiv 2020, arXiv:2009.05977.
- Lei, B.; Xia, Z.; Jiang, F.; Jiang, X.; Ge, Z.; Xu, Y.; Qin, J.; Chen, S.; Wang, T.; Wang, S. Skin Lesion Segmentation via Generative Adversarial Networks with Dual Discriminators. Med. Image Anal. 2020, 64, 101716.
- International Skin Imaging Collaboration (ISIC) Sixth ISIC Skin Image Analysis CVPR 2021 Virtual. Available online: https://workshop2021.isic-archive.com (accessed on 15 October 2022).
- Winkler, J.K.; Sies, K.; Fink, C.; Toberer, F.; Enk, A.; Deinlein, T.; Hofmann-Wellenhof, R.; Thomas, L.; Lallas, A.; Blum, A.; et al. Melanoma Recognition by a Deep Learning Convolutional Neural Network—Performance in Different Melanoma Subtypes and Localisations. Eur. J. Cancer 2020, 127, 21–29.
- Ferrara, G.; Argenziano, G. The WHO 2018 Classification of Cutaneous Melanocytic Neoplasms: Suggestions from Routine Practice. Front. Oncol. 2021, 11, 675296.
- Braun, R.P.; Rabinovitz, H.S.; Oliviero, M.; Kopf, A.W.; Saurat, J.-H. Dermoscopy of Pigmented Skin Lesions. J. Am. Acad. Dermatol. 2005, 52, 109–121.
- Binder, M.; Steiner, A.; Schwarz, M.; Knollmayer, S.; Wolff, K.; Pehamberger, H. Application of an Artificial Neural Network in Epiluminescence Microscopy Pattern Analysis of Pigmented Skin Lesions: A Pilot Study. Br. J. Dermatol. 1994, 130, 460–465.
- Sies, K.; Winkler, J.K.; Fink, C.; Bardehle, F.; Toberer, F.; Buhl, T.; Enk, A.; Blum, A.; Rosenberger, A.; Haenssle, H.A. Past and Present of Computer-Assisted Dermoscopic Diagnosis: Performance of a Conventional Image Analyser versus a Convolutional Neural Network in a Prospective Data Set of 1,981 Skin Lesions. Eur. J. Cancer 2020, 135, 39–46.
- Yang, Y.; Ge, Y.; Guo, L.; Wu, Q.; Peng, L.; Zhang, E.; Xie, J.; Li, Y.; Lin, T. Development and Validation of Two Artificial Intelligence Models for Diagnosing Benign, Pigmented Facial Skin Lesions. Ski. Res. Technol. 2021, 27, 74–79.
- Tschandl, P.; Codella, N.; Akay, B.N.; Argenziano, G.; Braun, R.P.; Cabo, H.; Gutman, D.; Halpern, A.; Helba, B.; Hofmann-wellenhof, R.; et al. Comparison of the Accuracy of Human Readers versus Machine-Learning Algorithms for Pigmented Skin Lesion Classification: An Open, Web-Based, International, Diagnostic Study. Lancet Oncol. 2019, 20, 938–947.
- Lyakhov, P.A.; Lyakhova, U.A.; Nagornov, N.N. System for the Recognizing of Pigmented Skin Lesions with Fusion and Analysis of Heterogeneous Data Based on a Multimodal Neural Network. Cancers 2022, 14, 1819.
- Penn, L.; Rothman, L.; Sutton, A.M.; Brinster, N.K.; Vidal, C.I. What’s New in Dermatopathology: Inflammatory Dermatoses. Adv. Anat. Pathol. 2019, 26, 40–55.
- De Guzman, L.C.; Maglaque, R.P.C.; Torres, V.M.B.; Zapido, S.P.A.; Cordel, M.O. Design and Evaluation of a Multi-Model, Multi-Level Artificial Neural Network for Eczema Skin Lesion Detection. In Proceedings of the 2015 3rd International Conference on Artificial Intelligence, Modelling and Simulation (AIMS), Kota Kinabalu, Malaysia, 2–4 December 2015; pp. 42–47.
- Shen, X.; Zhang, J.; Yan, C.; Zhou, H. An Automatic Diagnosis Method of Facial Acne Vulgaris Based on Convolutional Neural Network. Sci. Rep. 2018, 8, 5839.
- Seité, S.; Khammari, A.; Benzaquen, M.; Moyal, D.; Dréno, B. Development and Accuracy of an Artificial Intelligence Algorithm for Acne Grading from Smartphone Photographs. Exp. Dermatol. 2019, 28, 1252–1257.
- Han, S.S.; Park, G.H.; Lim, W.; Kim, M.S.; Na, J.I.; Park, I.; Chang, S.E. Deep Neural Networks Show an Equivalent and Often Superior Performance to Dermatologists in Onychomycosis Diagnosis: Automatic Construction of Onychomycosis Datasets by Region-Based Convolutional Deep Neural Network. PLoS ONE 2018, 13, e0191493.
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the Global Cancer Incidence and Mortality in 2018: GLOBOCAN Sources and Methods. Int. J. Cancer 2019, 144, 1941–1953.
- Leiter, U.; Keim, U.; Garbe, C. Epidemiology of Skin Cancer: Update 2019. In Sunlight, Vitamin D and Skin Cancer; Reichrath, J., Ed.; Springer: Cham, Switzerland, 2020; ISBN 9780123819789.
- Lomas, A.; Leonardi-Bee, J.; Bath-Hextall, F. A Systematic Review of Worldwide Incidence of Nonmelanoma Skin Cancer. Br. J. Dermatol. 2012, 166, 1069–1080.
- Schadendorf, D.; van Akkooi, A.C.J.; Berking, C.; Griewank, K.G.; Gutzmer, R.; Hauschild, A.; Stang, A.; Roesch, A.; Ugurel, S. Melanoma. Lancet 2018, 392, 971–984.
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249.
- Friedman, R.J.; Rigel, D.S.; Kopf, A.W. Early Detection of Malignant Melanoma: The Role of Physician Examination and Self-Examination of the Skin. CA Cancer J. Clin. 1985, 35, 130–151.
- Magro, C.M.; Neil Crowson, A.; Mihm, M.C. Unusual Variants of Malignant Melanoma. Mod. Pathol. 2006, 19, 41–70.
- Blum, A.; Luedtke, H.; Ellwanger, U.; Schwabe, R.; Rassner, G.; Garbe, C. Digital Image Analysis for Diagnosis of Cutaneous Melanoma. Development of a Highly Effective Computer Algorithm Based on Analysis of 837 Melanocytic Lesions. Br. J. Dermatol. 2004, 151, 1029–1038.
- Marchetti, M.A.; Codella, N.C.F.; Dusza, S.W.; Gutman, D.A.; Helba, B.; Kalloo, A.; Mishra, N.; Carrera, C.; Celebi, M.E.; DeFazio, J.L.; et al. Results of the 2016 International Skin Imaging Collaboration International Symposium on Biomedical Imaging Challenge: Comparison of the Accuracy of Computer Algorithms to Dermatologists for the Diagnosis of Melanoma from Dermoscopic Images. J. Am. Acad. Dermatol. 2018, 78, 270–277.e1.
- Marchetti, M.A.; Liopyris, K.; Dusza, S.W.; Codella, N.C.F.; Gutman, D.A.; Helba, B.; Kalloo, A.; Halpern, A.C. International Skin Imaging Collaboration. Computer algorithms show potential for improving dermatologists’ accuracy to diagnose cutaneous melanoma: Results of the International Skin Imaging Collaboration 2017. J. Am. Acad. Dermatol. 2020, 82, 622–627.
- Tang, P.; Liang, Q.; Yan, X.; Xiang, S.; Sun, W.; Zhang, D.; Coppola, G. Efficient Skin Lesion Segmentation Using Separable-Unet with Stochastic Weight Averaging. Comput. Methods Programs Biomed. 2019, 178, 289–301.
- MacLellan, A.N.; Price, E.L.; Publicover-Brouwer, P.; Matheson, K.; Ly, T.Y.; Pasternak, S.; Walsh, N.M.; Gallant, C.J.; Oakley, A.; Hull, P.R.; et al. The Use of Non-Invasive Imaging Techniques in the Diagnosis of Melanoma: A Prospective Diagnostic Accuracy Study. J. Am. Acad. Dermatol. 2020, 85, 353–359.
- Winkler, J.K.; Sies, K.; Fink, C.; Toberer, F.; Enk, A.; Abassi, M.S.; Fuchs, T.; Haenssle, H.A. Association between Different Scale Bars in Dermoscopic Images and Diagnostic Performance of a Market-Approved Deep Learning Convolutional Neural Network for Melanoma Recognition. Eur. J. Cancer 2021, 145, 146–154.
- Sinclair, R.; Meah, N.; Arasu, A. Skin Checks in Primary Care. Aust. J. Gen. Pract. 2019, 48, 614–619.
- Rayner, J.E.; Laino, A.M.; Nufer, K.L.; Adams, L.; Raphael, A.P.; Menzies, S.W.; Soyer, H.P. Clinical Perspective of 3D Total Body Photography for Early Detection and Screening of Melanoma. Front. Med. 2018, 5, 152.
- Primiero, C.A.; McInerney-Leo, A.M.; Betz-Stablein, B.; Whiteman, D.C.; Gordon, L.; Caffery, L.; Aitken, J.F.; Eakin, E.; Osborne, S.; Gray, L.; et al. Evaluation of the Efficacy of 3D Total-Body Photography with Sequential Digital Dermoscopy in a High-Risk Melanoma Cohort: Protocol for a Randomised Controlled Trial. BMJ Open 2019, 9, e032969.
- Roffman, D.; Hart, G.; Girardi, M.; Ko, C.J.; Deng, J. Predicting Non-Melanoma Skin Cancer via a Multi-Parameterized Artificial Neural Network. Sci. Rep. 2018, 8, 1701.
- Alzubaidi, L.; Al-Amidie, M.; Al-Asadi, A.; Humaidi, A.J.; Al-Shamma, O.; Fadhel, M.A.; Zhang, J.; Santamaría, J.; Duan, Y. Novel Transfer Learning Approach for Medical Imaging with Limited Labeled Data. Cancers 2021, 13, 1590.
- McClatchey, A.I. Neurofibromatosis. Annu. Rev. Pathol. 2007, 2, 191–216.
- Boyd, K.P.; Korf, B.R.; Theos, A. Neurofibromatosis Type 1. J. Am. Acad. Dermatol. 2009, 61, 1–14; quiz 15–16, quiz 15–16.
- Wei, C.J.; Yan, C.; Tang, Y.; Wang, W.; Gu, Y.H.; Ren, J.Y.; Cui, X.W.; Lian, X.; Liu, J.; Wang, H.J.; et al. Computed Tomography–Based Differentiation of Benign and Malignant Craniofacial Lesions in Neurofibromatosis Type I Patients: A Machine Learning Approach. Front. Oncol. 2020, 10, 1192.
- Ho, C.Y.; Kindler, J.M.; Persohn, S.; Kralik, S.F.; Robertson, K.A.; Territo, P.R. Image Segmentation of Plexiform Neurofibromas from a Deep Neural Network Using Multiple B-Value Diffusion Data. Sci. Rep. 2020, 10, 17857.
- Bashat, D.B.; Artzi, M.; Ganut, T.; Vitinshtein, F.; Ben-Sira, L.; Bokstein, F. Differentiation between Plexiform Neurofibromas and Malignant Nerve Sheath Tumors in Patients with Neurofibromatosis Type 1 (NF1) Using Radiomics Analysis of MRI. In Proceedings of the European Congress of Radiology-ECR 2019, Vienna, Austria, 27 February–3 March 2019.
- Duarte, J.V.; Ribeiro, M.J.; Violante, I.R.; Cunha, G.; Silva, E.; Castelo-Branco, M. Multivariate Pattern Analysis Reveals Subtle Brain Anomalies Relevant to the Cognitive Phenotype in Neurofibromatosis Type 1. Hum. Brain Mapp. 2014, 35, 89–106.
- Parisi, R.; Symmons, D.P.M.; Griffiths, C.E.M.; Ashcroft, D.M. Global Epidemiology of Psoriasis: A Systematic Review of Incidence and Prevalence. J. Investig. Dermatol. 2013, 133, 377–385.
- van de Kerkhof, P.C. the Psoriasis Area and Severity Index and Alternative Approaches for the Assessment of Severity: Persisting Areas of Confusion. Br. J. Dermatol. 1997, 137, 661–662.
- Walsh, J.A.; McFadden, M.; Woodcock, J.; Clegg, D.O.; Helliwell, P.; Dommasch, E.; Gelfand, J.M.; Krueger, G.G.; Duffin, K.C. Product of the Physician Global Assessment and Body Surface Area: A Simple Static Measure of Psoriasis Severity in a Longitudinal Cohort. J. Am. Acad. Dermatol. 2013, 69, 931–937.
- Bozek, A.; Reich, A. The Reliability of Three Psoriasis Assessment Tools: Psoriasis Area and Severity Index, Body Surface Area and Physician Global Assessment. Adv. Clin. Exp. Med. 2017, 26, 851–856.
- Meienberger, N.; Anzengruber, F.; Amruthalingam, L.; Christen, R.; Koller, T.; Maul, J.T.; Pouly, M.; Djamei, V.; Navarini, A.A. Observer-Independent Assessment of Psoriasis-Affected Area Using Machine Learning. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 1362–1368.
- Fink, C.; Fuchs, T.; Enk, A.; Haenssle, H.A. Design of an Algorithm for Automated, Computer-Guided PASI Measurements by Digital Image Analysis. J. Med. Syst. 2018, 42, 248.
- Fink, C.; Alt, C.; Uhlmann, L.; Klose, C.; Enk, A.; Haenssle, H.A. Precision and Reproducibility of Automated Computer-Guided Psoriasis Area and Severity Index Measurements in Comparison with Trained Physicians. Br. J. Dermatol. 2019, 180, 390–396.
- Pal, A.; Chaturvedi, A.; Garain, U.; Chandra, A.; Chatterjee, R. Severity Grading of Psoriatic Plaques Using Deep CNN Based Multi-Task Learning. In Proceedings of the 2016 23rd International Conference on Pattern Recognition (ICPR), Cancun, Mexico, 4–8 December 2016; pp. 1478–1483.
- Pal, A.; Chaturvedi, A.; Chandra, A.; Chatterjee, R.; Senapati, S.; Frangi, A.F.; Garain, U. MICaps: Multi-Instance Capsule Network for Machine Inspection of Munro’s Microabscess. Comput. Biol. Med. 2022, 140, 105071.
- Zhao, S.; Xie, B.; Li, Y.; Zhao, X.; Kuang, Y.; Su, J.; He, X.; Wu, X.; Fan, W.; Huang, K.; et al. Smart Identification of Psoriasis by Images Using Convolutional Neural Networks: A Case Study in China. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 518–524.
- Emam, S.; Du, A.X.; Surmanowicz, P.; Thomsen, S.F.; Greiner, R.; Gniadecki, R. Predicting the Long-Term Outcomes of Biologics in Patients with Psoriasis Using Machine Learning. Br. J. Dermatol. 2020, 182, 1305–1307.
- Diepgen, T.L.; Andersen, K.E.; Chosidow, O.; Coenraads, P.J.; Elsner, P.; English, J.; Fartasch, M.; Gimenez-Arnau, A.; Nixon, R.; Sasseville, D.; et al. Guidelines for Diagnosis, Prevention and Treatment of Hand Eczema—Short Version. JDDG—J. Ger. Soc. Dermatol. 2015, 13, 77–85.
- Han, S.S.; Park, I.; Eun Chang, S.; Lim, W.; Kim, M.S.; Park, G.H.; Chae, J.B.; Huh, C.H.; Na, J.I. Augmented Intelligence Dermatology: Deep Neural Networks Empower Medical Professionals in Diagnosing Skin Cancer and Predicting Treatment Options for 134 Skin Disorders. J. Investig. Dermatol. 2020, 140, 1753–1761.
- Schnürle, S.; Pouly, M.; Vor Der Brück, T.; Navarini, A.; Koller, T. On Using Support Vector Machines for the Detection and Quantification of Hand Eczema. In Proceedings of the 9th International Conference on Agents and Artificial Intelligence (ICAART 2017), Porto, Portugal, 24–26 February 2017; Volume 2, pp. 75–84.
- Garzorz-Stark, N.; Eyerich, K. Molecular Diagnostics of Hand Eczema. Hautarzt 2019, 70, 760–765.
- Weidinger, S.; Novak, N. Atopic Dermatitis. Lancet 2016, 387, 1109–1122.
- Drucker, A.M.; Wang, A.R.; Li, W.Q.; Sevetson, E.; Block, J.K.; Qureshi, A.A. The Burden of Atopic Dermatitis: Summary of a Report for the National Eczema Association. J. Investig. Dermatol. 2017, 137, 26–30.
- Gustafson, E.; Pacheco, J.; Wehbe, F.; Silverberg, J.; Thompson, W. A Machine Learning Algorithm for Identifying Atopic Dermatitis in Adults from Electronic Health Records. In Proceedings of the 2017 IEEE International Conference on Healthcare Informatics (ICHI), Park City, UT, USA, 23–26 August 2017; pp. 83–90.
- Patella, V.; Florio, G.; Palmieri, M.; Bousquet, J.; Tonacci, A.; Giuliano, A.; Gangemi, S. Atopic Dermatitis Severity during Exposure to Air Pollutants and Weather Changes with an Artificial Neural Network (ANN) Analysis. Pediatr. Allergy Immunol. 2020, 31, 938–945.
- Guimarães, P.; Batista, A.; Zieger, M.; Kaatz, M.; Koenig, K. Artificial Intelligence in Multiphoton Tomography: Atopic Dermatitis Diagnosis. Sci. Rep. 2020, 10, 7968.
- Melina, A.; Dinh, N.N.; Tafuri, B.; Schipani, G.; Nisticò, S.; Cosentino, C.; Amato, F.; Thiboutot, D.; Cherubini, A. Artificial Intelligence for the Objective Evaluation of Acne Investigator Global Assessment. J. Drugs Dermatol. 2018, 17, 1006–1009.
- Maul, L.V.; Meienberger, N.; Kaufmann, L. Role of Artificial Intelligence in Assessing the Extent and Progression of Dermatoses. Hautarzt 2020, 71, 677–685.
- Luo, W.; Liu, J.; Huang, Y.; Zhao, N. An Effective Vitiligo Intelligent Classification System. J. Ambient Intell. Humaniz. Comput. 2020.
- Low, M.; Huang, V.; Raina, P. Automating Vitiligo Skin Lesion Segmentation Using Convolutional Neural Networks. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1992–1995.
- Gao, W.; Li, M.; Wu, R.; Du, W.; Zhang, S.; Yin, S.; Chen, Z.; Huang, H. The Design and Application of an Automated Microscope Developed Based on Deep Learning for Fungal Detection in Dermatology. Mycoses 2021, 64, 245–251.
- Brewer, A.C.; Endly, D.C.; Henley, J.; Amir, M.; Sampson, B.P.; Moreau, J.F.; Dellavalle, R.P. Mobile Applications in Dermatology. JAMA Dermatol. 2013, 149, 1300–1304.
- De, A. Next-Generation Technologies in Dermatology: Use of Artificial Intelligence and Mobile Applications. Indian J. Dermatol. 2020, 65, 351.
- Zhang, L.; Zhang, D.; Sun, M.M.; Chen, F.M. Facial Beauty Analysis Based on Geometric Feature: Toward Attractiveness Assessment Application. Expert Syst. Appl. 2017, 82, 252–265.
- Swetter, S.M.; Tsao, H.; Bichakjian, C.K.; Curiel-Lewandrowski, C.; Elder, D.E.; Gershenwald, J.E.; Guild, V.; Grant-Kels, J.M.; Halpern, A.C.; Johnson, T.M.; et al. Guidelines of Care for the Management of Primary Cutaneous Melanoma. J. Am. Acad. Dermatol. 2019, 80, 208–250.
- Tintle, S.M.; Keeling, J.J.; Shawen, S.B.; Forsberg, J.A.; Potter, B.K. Traumatic and Trauma-Related Amputations: Part I: General Principles and Lower-Extremity Amputations. J. Bone Jt. Surg. Am. 2010, 92, 2852–2868.
- Tintle, S.M.; Baechler, M.F.; Nanos, G.P., 3rd; Forsberg, J.A.; Potter, B.K. Traumatic and Trauma-Related Amputations: Part II: Upper Extremity and Future Directions. J. Bone Jt. Surg. Am. 2010, 92, 2934–2945.
- Harwell, R.C.; Ferguson, R.L. Physiologic Tremor and Microsurgery. Microsurgery 1983, 4, 187–192.
- Bodenstedt, S.; Wagner, M.; Müller-Stich, B.P.; Weitz, J.; Speidel, S. Artificial Intelligence-Assisted Surgery: Potential and Challenges. Visc. Med. 2020, 36, 450–455.
- Fagius, J.; Nordin, M.; Wall, M. Sympathetic Nerve Activity to Amputated Lower Leg in Humans: Evidence of Altered Skin Vasoconstrictor Discharge. Pain 2002, 98, 37–45.
- Cutrone, A.; Micera, S. Implantable Neural Interfaces and Wearable Tactile Systems for Bidirectional Neuroprosthetics Systems. Adv. Healthc. Mater. 2019, 8, e1801345.
- Parajuli, N.; Sreenivasan, N.; Bifulco, P.; Cesarelli, M.; Savino, S.; Niola, V.; Esposito, D.; Hamilton, T.J.; Naik, G.R.; Gunawardana, U.; et al. Real-Time EMG Based Pattern Recognition Control for Hand Prostheses: A Review on Existing Methods, Challenges and Future Implementation. Sensors 2019, 19, 4596.
- Sohn, W.; Finley, D.S.; Jakowatz, J.; Ornstein, D.K. Robot-Assisted Laparoscopic Transperitoneal Pelvic Lymphadenectomy and Metastasectomy for Melanoma: Initial Report of Two Cases. J. Robot. Surg. 2010, 4, 129–132.
- Kim, S.I.; Lee, S.; Jeong, C.W.; Kim, H.S. Robot-Assisted Anterior Pelvic Exenteration in Vulvovaginal Malignant Melanoma. Gynecol. Oncol. 2018, 148, 430–431.
- Hyde, G.A.; Jung, N.L.; Valle, A.A.; Bhattacharya, S.D.; Keel, C.E. Robotic Inguinal Lymph Node Dissection for Melanoma: A Novel Approach to a Complicated Problem. J. Robot. Surg. 2018, 12, 745–748.
- Miura, J.T.; Dossett, L.A.; Thapa, R.; Kim, Y.; Potdar, A.; Daou, H.; Sun, J.; Sarnaik, A.A.; Zager, J.S. Robotic-Assisted Pelvic Lymphadenectomy for Metastatic Melanoma Results in Durable Oncologic Outcomes. Ann. Surg. Oncol. 2020, 27, 196–202.
- Rose, P.T.; Nusbaum, B. Robotic Hair Restoration. Dermatol. Clin. 2014, 32, 97–107.
- Bicknell, L.M.; Kash, N.; Kavouspour, C.; Rashid, R.M. Follicular Unit Extraction Hair Transplant Harvest: A Review of Current Recommendations and Future Considerations. Dermatol. Online J. 2014, 20, doj_21754.
More
