Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Conner Chen and Version 1 by ALESSANDRO Giammona.
Hedgehog–GLI (HH) signaling plays an essential role in embryogenesis and tissue homeostasis. Aberrant activation of the pathway through mutations or other mechanisms is involved in the development and progression of numerous types of cancer, including basal cell carcinoma, medulloblastoma, melanoma, breast, prostate, hepatocellular and pancreatic carcinomas. Activation of HH signaling sustains proliferation, suppresses cell death signals, enhances invasion and metastasis, deregulates cellular metabolism and promotes angiogenesis and tumor inflammation. Targeted inhibition of the HH pathway has therefore emerged as an attractive therapeutic strategy for the treatment of a wide range of cancers.
- hedgehog signaling
- tumor microenvironment
- immunosuppression
1. Introduction
The Hedgehog–GLI (HH) signaling pathway plays key roles during embryonic development and is involved in cell proliferation, differentiation and tissue patterning. In adults, HH signaling is rapidly turned off and remains active in the stem cells of the central nervous system, skin and intestine, where it maintains tissue homeostasis and regeneration [1]. The HH signaling is aberrantly activated during the initiation and progression of a variety of cancer types, including those of the brain, skin, breast, prostate, hepatocellular and pancreatic carcinomas and hematological malignancies. The HH pathway is involved in enhancing proliferation, invasion and metastasis, in suppressing cell death signals and in deregulating the cellular metabolism [2]. Several reports have implicated HH signaling in suppressing the immune system and promoting an immunosuppressive tumor microenvironment (TME) [3]. Recent advances in cancer immunology and immunotherapy have emphasized the need for an accurate understanding of the immune-modulatory functions of oncogenic signaling pathways and their role in cancer immunity.
2. Hedgehog Signaling Pathway
The HH pathway is an evolutionary signaling pathway that plays a pivotal role in patterning and organogenesis during embryonic development and in adult tissue homeostasis and repair [1]. This complex transduction pathway is coordinated by several regulatory components and post-translational modifications. In mammals, HH signaling consists of three secreted HH ligands (Sonic Hedgehog, SHH; Desert Hedgehog, DHH; and Indian Hedgehog, IHH); the 12-pass transmembrane receptor Patched 1 (PTCH1); the 7-pass transmembrane G protein-coupled receptor (GPCR) Smoothened (SMO), as the main transducer of the HH pathway; and the three zinc finger GLI transcription factors (GLI1, GLI2, GLI3), as the final mediators of the transcriptional response of HH signaling [4]. Additional members include a number of regulatory kinases [5] and Suppressor of Fused (SUFU), the main negative regulator of the GLI [6] (Figure 1).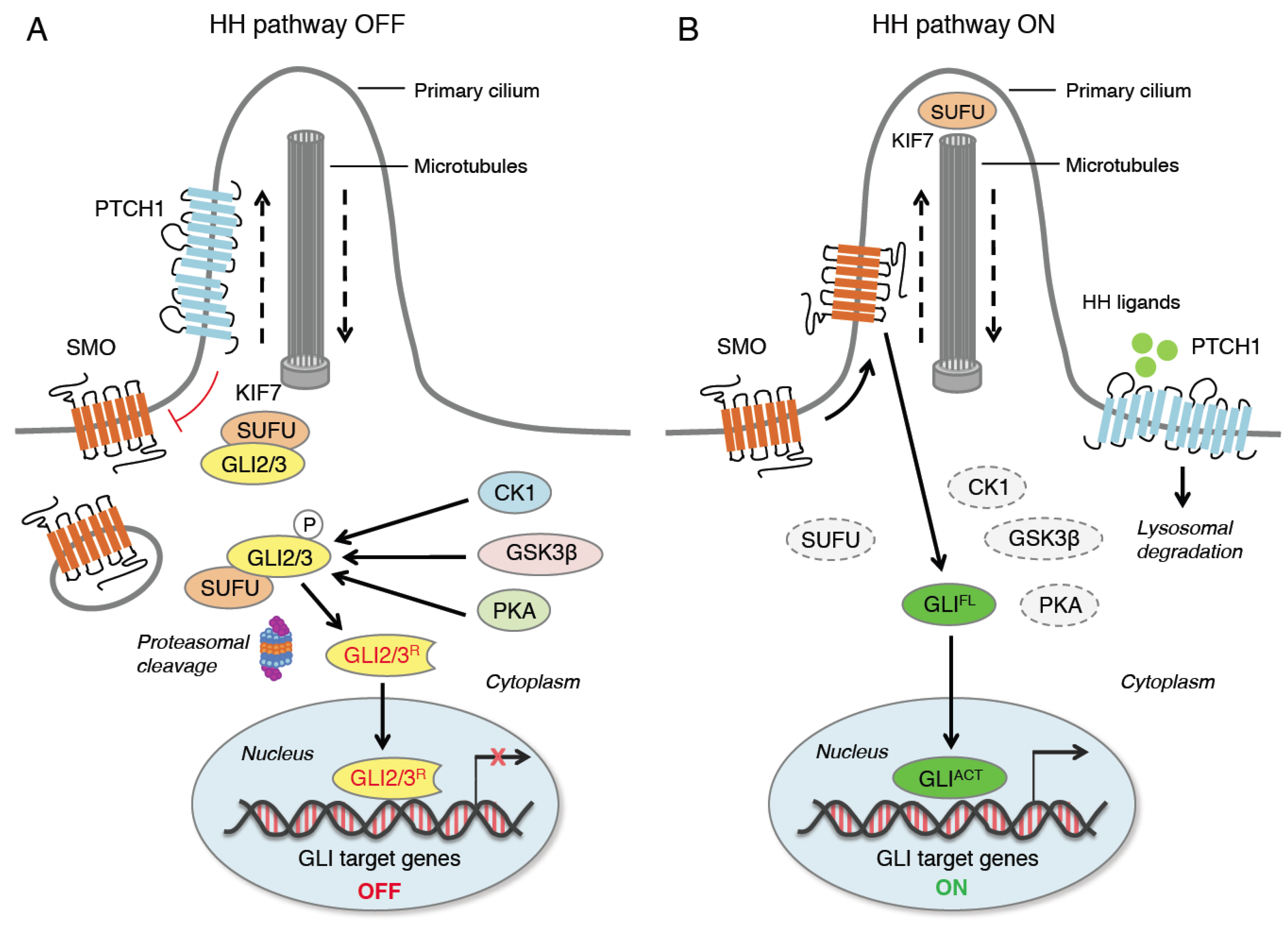
Figure 1. Canonical activation of HH signaling. When HH ligands are not present (A), PTCH1 represses SMO by preventing its entry into the primary cilium (PC). GLI2 and GLI3 are sequestered in the cytoplasm by SUFU and phosphorylated by PKA, CK1 and GSK3β. The GLI undergo ubiquitination through the E3 ubiquitin ligase β-TrCP. GLI1 is fully degraded, whereas GLI3 and, to a lesser extent, GLI2 undergo partial proteasome degradation, leading to the formation of repressor forms (GLI3/2R) that move into the nucleus inhibiting the transcription of GLI target genes. In the presence of HH ligands (B), PTCH1 is displaced from the PC and undergoes lysosomal degradation, and SMO translocates into the PC. Active SMO relieves the SUFU-mediated suppression of GLI2 and GLI3, triggering a signaling cascade that leads to the translocation of full length activated forms of GLI (GLIACT) into the nucleus, where they promote the transcription of GLI target genes. KIF7 is a kinesin protein that acts in anterograde transport (from base to tip) of the PC. CK1, casein kinase 1; GLI2/3R, GLI2/3 repressors; GLIACT, GLI activators; GLIFL, GLI full length; GSK3β, glycogen synthase kinase 3β; HH, Hedgehog; KIF7, kinesin family member 7; PKA, protein kinase A; PTCH1, Patched 1; SMO, Smoothened; SUFU, Suppressor of Fused; β-TrCP, β-transducin repeat-containing protein.
References
- Ingham, P.W.; McMahon, A.P. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001, 15, 3059–3087.
- Hanna, A.; Shevde, L.A. Hedgehog signaling: Modulation of cancer properties and tumor microenvironment. Mol. Cancer 2016, 15, 24.
- Grund-Gröschke, S.; Stockmaier, G.; Aberger, F. Hedgehog/GLI signaling in tumor immunity—New therapeutic opportunities and clinical implications. Cell Commun. Signal. 2019, 17, 172.
- Varjosalo, M.; Taipale, J. Hedgehog: Functions and mechanisms. Genes Dev. 2008, 22, 2454–2472.
- Montagnani, V.; Stecca, B. Role of Protein Kinases in Hedgehog Pathway Control and Implications for Cancer Therapy. Cancers 2019, 11, 449.
- Svärd, J.; Heby-Henricson, K.; Persson-Lek, M.; Rozell, B.; Lauth, M.; Bergström, A.; Ericson, J.; Toftgård, R.; Teglund, S. Genetic elimination of suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev. Cell 2006, 10, 187–197.
- Goetz, S.C.; Anderson, K.V. The primary cilium: A signalling centre during vertebrate development. Nat. Rev. Genet. 2010, 11, 331–344.
- Kogerman, P.; Grimm, T.; Kogerman, L.; Krause, D.; Undén, A.B.; Sandstedt, B.; Toftgård, R.; Zaphiropoulos, P.G. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell Biol. 1999, 1, 312–319.
- Wang, B.; Li, Y. Evidence for the direct involvement of βTrCP in Gli3 protein processing. Proc. Natl. Acad. Sci. USA 2006, 103, 33–38.
- Pan, Y.; Wang, B. A novel protein-processing domain in Gli2 and Gli3 differentially blocks complete protein degradation by the proteasome. J. Biol. Chem. 2007, 282, 10846–10852.
- Niewiadomski, P.; Kong, J.H.; Ahrends, R.; Ma, Y.; Humke, E.W.; Khan, S.; Teruel, M.N.; Novitch, B.G.; Rohatgi, R. Gli protein activity is controlled by multisite phosphorylation in vertebrate Hedgehog signaling. Cell Rep. 2014, 6, 168–181.
- Pan, Y.; Bai, C.B.; Joyner, A.L.; Wang, B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol. Cell. Biol. 2006, 26, 3365–3377.
- Pepelyayeva, Y.; Amalfitano, A. The role of ERAP1 in autoinflammation and autoimmunity. Hum. Immunol. 2019, 80, 302–309.
- Bufalieri, F.; Infante, P.; Bernardi, F.; Caimano, M.; Romania, P.; Moretti, M.; Lospinoso Severini, L.; Talbot, J.; Melaiu, O.; Tanori, M.; et al. ERAP1 promotes Hedgehog-dependent tumorigenesis by controlling USP47-mediated degradation of βTrCP. Nat. Commun. 2019, 10, 3304.
- Rohatgi, R.; Milenkovic, L.; Scott, M.P. Patched1 regulates hedgehog signaling at the primary cilium. Science 2007, 317, 372–376.
- Katoh, Y.; Katoh, M. Hedgehog target genes: Mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr. Mol. Med. 2009, 9, 873–886.
- Sigafoos, A.N.; Paradise, B.D.; Fernandez-Zapico, M.E. Hedgehog/GLI Signaling Pathway: Transduction, Regulation, and Implications for Disease. Cancers 2021, 13, 3410.
- Pietrobono, S.; Gagliardi, S.; Stecca, B. Non-Canonical Hedgehog Signaling Pathway in Cancer: Activation of GLI Transcription Factors Beyond Smoothened. Front. Genet. 2019, 10, 556.
- Riobo, N.A.; Haines, G.M.; Emerson, C.P., Jr. Protein kinase C-delta and mitogen-activated protein/extracellular signal-regulated kinase-1 control GLI activation in hedgehog signaling. Cancer Res. 2006, 66, 839–845.
- Riobó, N.A.; Lu, K.; Ai, X.; Haines, G.M.; Emerson, C.P., Jr. Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 4505–4510.
- Stecca, B.; Mas, C.; Clement, V.; Zbinden, M.; Correa, R.; Piguet, V.; Beermann, F.; Ruiz i Altaba, A. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc. Natl. Acad. Sci. USA 2007, 104, 5895–5900.
- Wang, Y.; Ding, Q.; Yen, C.J.; Xia, W.; Izzo, J.G.; Lang, J.Y.; Li, C.W.; Hsu, J.L.; Miller, S.A.; Wang, X.; et al. The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer Cell 2012, 21, 374–387.
- Dennler, S.; André, J.; Alexaki, I.; Li, A.; Magnaldo, T.; ten Dijke, P.; Wang, X.J.; Verrecchia, F.; Mauviel, A. Induction of sonic hedgehog mediators by transforming growth factor-beta: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res. 2007, 67, 6981–6986.
- Atwood, S.X.; Li, M.; Lee, A.; Tang, J.Y.; Oro, A.E. GLI activation by atypical protein kinase C ι/λ regulates the growth of basal cell carcinomas. Nature 2013, 494, 484–488.
- Zwerner, J.P.; Joo, J.; Warner, K.L.; Christensen, L.; Hu-Lieskovan, S.; Triche, T.J.; May, W.A. The EWS/FLI1 oncogenic transcription factor deregulates GLI1. Oncogene 2008, 27, 3282–3291.
- Stecca, B.; Ruiz i Altaba, A. A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. EMBO J. 2009, 28, 663–676.
- Jagani, Z.; Mora-Blanco, E.L.; Sansam, C.G.; McKenna, E.S.; Wilson, B.; Chen, D.; Klekota, J.; Tamayo, P.; Nguyen, P.T.; Tolstorukov, M.; et al. Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nat. Med. 2010, 16, 1429–1433.
- Tang, Y.; Gholamin, S.; Schubert, S.; Willardson, M.I.; Lee, A.; Bandopadhayay, P.; Bergthold, G.; Masoud, S.; Nguyen, B.; Vue, N.; et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat. Med. 2014, 20, 732–740.
- Canettieri, G.; Di Marcotullio, L.; Greco, A.; Coni, S.; Antonucci, L.; Infante, P.; Pietrosanti, L.; De Smaele, E.; Ferretti, E.; Miele, E.; et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat. Cell Biol. 2010, 12, 132–142.
- Jiang, J.; Hui, C.C. Hedgehog signaling in development and cancer. Dev. Cell 2008, 15, 801–812.
- McMillan, R.; Matsui, W. Molecular pathways: The hedgehog signaling pathway in cancer. Clin. Cancer Res. 2012, 18, 4883–4888.
- Amakye, D.; Jagani, Z.; Dorsch, M. Unraveling the therapeutic potential of the hedgehog pathway in cancer. Nat. Med. 2013, 19, 1410–1422.
- Infante, P.; Alfonsi, R.; Botta, B.; Mori, M.; Di Marcotullio, L. Targeting GLI factors to inhibit the Hedgehog pathway. Trends Pharm. Sci. 2015, 36, 547–558.
- Pietrobono, S.; Stecca, B. Targeting the Oncoprotein Smoothened by Small Molecules: Focus on Novel Acylguanidine Derivatives as Potent Smoothened Inhibitors. Cells 2018, 7, 272.
- Galperin, I.; Dempwolff, L.; Diederich, W.E.; Lauth, M. Inhibiting Hedgehog: An Update on Pharmacological Compounds and Targeting Strategies. J. Med. Chem. 2019, 62, 8392–8411.
- Gampala, S.; Yang, J.Y. Hedgehog Pathway Inhibitors against Tumor Microenvironment. Cells 2021, 10, 3135.
- Nguyen, N.M.; Cho, J. Hedgehog Pathway Inhibitors as Targeted Cancer Therapy and Strategies to Overcome Drug Resistance. Int. J. Mol. Sci. 2022, 23, 1733.
- LoRusso, P.M.; Rudin, C.M.; Reddy, J.C.; Tibes, R.; Weiss, G.J.; Borad, M.J.; Hann, C.L.; Brahmer, J.R.; Chang, I.; Darbonne, W.C.; et al. Phase I trial of hedgehog pathway inhibitor vismodegib (GDC-0449) in patients with refractory, locally advanced or metastatic solid tumors. Clin. Cancer Res. 2011, 17, 2502–2511.
- Von Hoff, D.D.; LoRusso, P.M.; Rudin, C.M.; Reddy, J.C.; Yauch, R.L.; Tibes, R.; Weiss, G.J.; Borad, M.J.; Hann, C.L.; Brahmer, J.R.; et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N. Engl. J. Med. 2009, 361, 1164–1172.
- Sekulic, A.; Migden, M.R.; Oro, A.E.; Dirix, L.; Lewis, K.D.; Hainsworth, J.D.; Solomon, J.A.; Yoo, S.; Arron, S.T.; Friedlander, P.A.; et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N. Engl. J. Med. 2012, 366, 2171–2179.
- Pan, S.; Wu, X.; Jiang, J.; Gao, W.; Wan, Y.; Cheng, D.; Han, D.; Liu, J.; Englund, N.P.; Wang, Y.; et al. Discovery of NVP-LDE225, a Potent and Selective Smoothened Antagonist. ACS Med. Chem. Lett. 2010, 1, 130–134.
- Munchhof, M.J.; Li, Q.; Shavnya, A.; Borzillo, G.V.; Boyden, T.L.; Jones, C.S.; LaGreca, S.D.; Martinez-Alsina, L.; Patel, N.; Pelletier, K.; et al. Discovery of PF-04449913, a Potent and Orally Bioavailable Inhibitor of Smoothened. ACS Med. Chem. Lett. 2011, 3, 106–111.
- Tremblay, M.R.; Lescarbeau, A.; Grogan, M.J.; Tan, E.; Lin, G.; Austad, B.C.; Yu, L.C.; Behnke, M.L.; Nair, S.J.; Hagel, M.; et al. Discovery of a potent and orally active hedgehog pathway antagonist (IPI-926). J. Med. Chem. 2009, 52, 4400–4418.
- Bender, M.H.; Hipskind, P.A.; Capen, A.R.; Cockman, M.; Credille, K.M.; Gao, H.; Bastian, J.A.; Clay, J.M.; Lobb, K.L.; Sall, D.J.; et al. Identification and characterization of a novel smoothened antagonist for the treatment of cancer with deregulated hedgehog signaling. Cancer Res. 2011, 71, 2819.
- Atwood, S.X.; Sarin, K.Y.; Whitson, R.J.; Li, J.R.; Kim, G.; Rezaee, M.; Ally, M.S.; Kim, J.; Yao, C.; Chang, A.L.; et al. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell 2015, 27, 342–353.
- Sharpe, H.J.; Pau, G.; Dijkgraaf, G.J.; Basset-Seguin, N.; Modrusan, Z.; Januario, T.; Tsui, V.; Durham, A.B.; Dlugosz, A.A.; Haverty, P.M.; et al. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell 2015, 27, 327–341.
- Lauth, M.; Bergström, A.; Shimokawa, T.; Toftgård, R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc. Natl. Acad. Sci. USA 2007, 104, 8455–8460.
- Infante, P.; Mori, M.; Alfonsi, R.; Ghirga, F.; Aiello, F.; Toscano, S.; Ingallina, C.; Siler, M.; Cucchi, D.; Po, A.; et al. Gli1/DNA interaction is a druggable target for Hedgehog-dependent tumors. EMBO J. 2015, 34, 200–217.
- Beauchamp, E.M.; Ringer, L.; Bulut, G.; Sajwan, K.P.; Hall, M.D.; Lee, Y.C.; Peaceman, D.; Ozdemirli, M.; Rodriguez, O.; Macdonald, T.J.; et al. Arsenic trioxide inhibits human cancer cell growth and tumor development in mice by blocking Hedgehog/GLI pathway. J. Clin. Investig. 2011, 121, 148–160.
- Kim, J.; Lee, J.J.; Kim, J.; Gardner, D.; Beachy, P.A. Arsenic antagonizes the Hedgehog pathway by preventing ciliary accumulation and reducing stability of the Gli2 transcriptional effector. Proc. Natl. Acad. Sci. USA 2010, 107, 13432–13437.
- Manetti, F.; Stecca, B.; Santini, R.; Maresca, L.; Giannini, G.; Taddei, M.; Petricci, E. Pharmacophore-Based Virtual Screening for Identification of Negative Modulators of GLI1 as Potential Anticancer Agents. ACS Med. Chem. Lett. 2020, 11, 832–838.
- Manetti, F.; Maresca, L.; Crivaro, E.; Pepe, S.; Cini, E.; Singh, S.; Governa, P.; Maramai, S.; Giannini, G.; Stecca, B.; et al. Quinolines and Oxazino-quinoline Derivatives as Small Molecule GLI1 Inhibitors Identified by Virtual Screening. ACS Med. Chem. Lett. 2022, 13, 1329–1336.
More
