Velvet deer are not only a representative special economic animal but also an important part of livestock. With the increasing awareness of international competition for germplasm resources in China, more and more attention has been paid to the protection and utilization of germplasm resources.
- velvet deer
- germplasm resources
- utilization
- China
1. Introduction
2. Present Situation of Velvet Deer Germplasm Resources
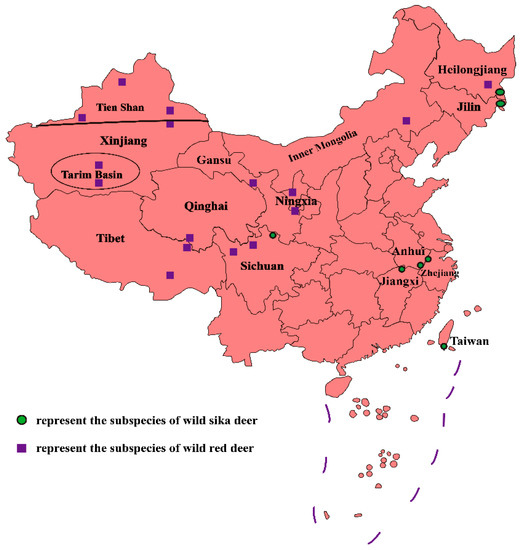
2.1. Present Situation of Wild Sika Deer Germplasm Resources
| Subspecies of Sika Deer | Distribution | Number |
|---|---|---|
| Cervus nippon hortulorum | Heilongjiang, Jilin province | --- |
| Cervus nippon kopshi | Qingliangfeng National Nature Reserve, Taohongling National Nature Reserve and south of Anhui | More than 874 |
| Cervus nippon sinchuanicus | Sichuan Tiebu Sike Deer Nature Reserve | About 800 |
| Cervus nippon taiouanus | Kenting National Park, Taiwan | About 1000 |
2.2. Present Situation of Wild Red Deer Germplasm Resources
| Subspecies of Red Deer | Distribution | Number |
|---|---|---|
| Cervus elaphus wallichi [11] | Sangri County, Tibet | About 300 |
| Cervus elaphus macneilli [1] | Aba and Ganzi, Sichuan Province | --- |
| Cervus elaphus kansuensis [12] | Qilian Mountain, Gansu, Qinhai, Ningxia, Southwest Sichuan and East Tibet | --- |
| Cervus elaphus yarkandensis [13] | Northern Tarim Basin and desert of the southern plain, Xinjiang | About 450 |
| Cervus elaphus songaricus [14] | Urumqi Nanshan, Hami Mountain and northern Tianshan Mountains, Xinjiang | Fewer than 10,000 |
| Cervus elaphus alashanicus [15] | Helan Mountain, Ningxia | --- |
| Cervus elaphus sibiricus [16] | Western and northern Xinjiang, Tianshan Mountain and forest and grassland in Altay | About 30,000 |
| Cervus elaphus xanthopygus [17][18][17,18] | Saihanwula Nature Reserve, Inner Mongolia and Muling forest area, Heilongjiang | --- |
2.3. Current Situation of Domestic Sika Deer Germplasm Resources
2.4. Current Situation of Domestic Red Deer Germplasm Resources
2.5. Current Situation of Other Kinds of Velvet Deer Germplasm Resources
3. Utilization of Velvet Deer Germplasm Resources
Germplasm resources are the material basis of modern breeding. Different types of germplasm resources have different genetic characteristics. An in-depth study of velvet deer germplasm resources will not only help to clarify the origin, evolution, classification and other issues, but it will also lay a great foundation for breeding innovation.
3.1. Application of Velvet Deer Germplasm Resources in Traditional Breeding
3.1.1. Pure Breeding
As a basic breeding method, pure breeding plays a role in maintaining and improving the good quality of varieties. It has been widely used in the breeding of pigs [30][30], sheep [31], cattle [32] and other domestic animals. Shuangyang sika deer are the first velvet-used variety in China. They have been selected and bred through large-scale pure breeding for 23 years and have been introduced around China [33]. In the 1960s, the average velvet antler yield of Shuangyang sika deer exceeded 1 kg, ranking first in antler production in China [34].
3.1.2. Cross-Breeding
Nowadays, antler velvet is a major source of income for the Chinese velvet deer industry. Therefore, velvet antler production performance has become the main goal of velvet deer breeding. As an important breeding method, hybridization is widely used in the modern livestock breeding industry and aims to breed new varieties with high production performance and improve the production efficiency of livestock, thereby achieving improved industrial gains. In order to improve the velvet antler-producing performance of velvet deer, China has carried out more than 70 years of cross-breeding with nearly 30 kinds of cross combinations, including hybridizations between red deer subspecies, sika deer varieties and hybridizations of sika deer and red deer [35].
Because of reproductive isolation, the offspring of most interspecific hybrids are not as adaptable as their parents. They are sterile or even lethal [36]. However, there is barely any hybridization incompatibility among velvet deer interspecies, which provides a feasible basis for variety improvement and breeding innovation of velvet deer. The characteristics of sika deer are relatively stable genetic performance, and the quality of their velvet antlers is better than that of red deer [37]. Sika deer are small in size and low in pilose antler yield, while red deer are large in size and high in pilose antler yield. Therefore, hybridization between sika deer and red deer has always been the main breeding method to improve production performance and breed new varieties. The physical appearance of F1s (Figure 2C,D) lies between the sika deer (Figure 2B) and the red deer (Figure 2A). F1s have normal reproductive capacity, the fertilityand the performance of antler-producing reflect the great economic heterosis [38]. In addition, the growth performance of F1s, such as dressing percentage, carcass weight and net meat weight [39], show extremely significant heterosis [40]. The cytogenetic basis for the fertility of interspecific hybridization in velvet deer is the formation of a cis-trivalent structure of the telocentric/centromere chromosome during the meiosis of F1, which may be related to the balanced distribution of gametes [41]. The testis sections of F1s also show normal morphology and viable sperm; the testes are fully developed [42]. The fertility of the interspecific hybridization greatly improves the breeding innovation efficiency of the velvet deer.

Figure 2.
Appearance of F1s and their parents: (
A
) male red deer and (
B
) female sika deer; (
C
,
D
) F1 offspring.
3.2. Application of Velvet Deer Germplasm Resources in Molecular Breeding
3.2.1. Molecular Assisted Breeding
With the development of biotechnology, the breeding of velvet deer has gradually developed to the molecular level. More and more molecular methods have been used to develop the utilization of velvet deer germplasm resources. The premise of the breeding innovation of velvet deer is to ensure the purity of the germplasm resources. The 1K chip, developed based on resequencing technology, can accurately distinguish sika deer, red deer and hybrids and judge the degree of hybridization [43]. Similarly, expression sequence labeling–simple sequence repeat (EST-SSR) is also used for identifying hybrids [44]. Based on the chip and EST-SSR, the parent individuals for breeding innovations can be scientifically selected. In addition, the unclear genetic relationships of velvet deer were also obstacles in the process of breeding innovation. Microsatellite markers are considered to be the first choice for characterizing the genetic diversity of populations [45], and they have been widely used for identifying the genetic relationship of species [46]. Ten STRs with high polymorphism have been developed to identify the genetic relationship among sika deer [47].
Molecular breeding of velvet deer is mainly based on molecular markers. Previous studies have found that ANXA2 [48], TRPC6 [49], HMG20A [49], and MTNR1A [50] are associated with velvet antler production performance. They also may become the candidate genes for screening paternal individuals during breeding. Transcriptome-based EST microsatellite markers have also been developed as important DNA markers for the selection of velvet antler producing performance [49][51].Molecular breeding of velvet deer is mainly based on molecular markers. Previous studies have found that ANXA2 [48], TRPC6 [49], HMG20A [49], and MTNR1A [50] are associated with velvet antler production performance. They also may become the candidate genes for screening paternal individuals during breeding. Transcriptome-based EST microsatellite markers have also been developed as important DNA markers for the selection of velvet antler producing performance [49,51].
3.2.2. Genomic Selection
Genomic selection (GS) is a molecular breeding technology that uses the whole genome’s genetic markers of reference groups with genotypes and phenotypes to build models and predict the phenotypic value of genotype-only individuals [52]. It has become an important method for improving the economic traits of livestock and poultry due to its high accuracy, good character-selection effect and reductions in the economic and time costs of breeding. The application of genomic selection cannot be separated from the popularization of whole-genome-sequencing technology. Obtaining a high-quality whole genome of a species is the prerequisite for genomic selection. The high-quality chromosome-level genome of sika deer has been published [53] and used as a reference genome for a genome-wide association study(GWAS). A total of 94 SNPs significantly related to velvet antler production performance have been obtained through genomic selection; they are located in the exon regions of OAS2 and ALYREF/THOC4 [54], and it is speculated that OAS2 and ALYREF/THOC4 may be related to the weight of velvet antlers [55][55]. As a member of Cervidae with special evolutionary status, the chromosome-level genome of Tarim red deer has also been published [56]. However, it was mainly used for study on adaptive evolution. There are no reports on genomic selection and molecular breeding of red deer.
