Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Camila Xu and Version 1 by Nimisha Sharma.
Most tree fruits are commercially grown on different root systems, hence called composite plants. The section provides the root system as the rootstock, and the atop ground portion is called the scion. The combination is selected based on different traits of scion varieties, rootstock, and prevailing edaphic situations. Rootstock selection is one of the most important factors in orchard management because it affects the growth, nutrient accumulation, environmental tolerance, and fruit quality of scion varieties.
- graft-union
- hormones
- molecular breeding
- rootstock
1. Introduction
Horticulture is currently a major potential sector for increasing agricultural production, income generation, and nutritional security through export, employment generation, value addition, and diversification. In addition to new major challenges, low productivity per unit area continues a problem in most of the horticultural crops, with climate change impacting the greater effect on fruit productivity. Biotic (causal agents of disease, insect, nematode, etc.), and abiotic (temperature, humidity, drought, wind, water logging, salinity, etc.), stresses are also challenges. Numerous studies on graft union formation and graft compatibility between scion–rootstock plants have given rise to several scientific hypotheses in herbaceous plants. However, due to long juvenile periods and generation times, and large plant size, studies at different growth stages of grafts in woody fruit plants are meager [1,2][1][2].
Rootstock selection is one of the most important factors in orchard management because it affects the growth, nutrient accumulation, environmental tolerance, and fruit quality of scion varieties [2,3,4][2][3][4]. Conventional breeding must be supplemented with molecular approaches to refine biotic and abiotic stress tolerance in fruit crops as lack of knowledge, breeding programs will often be time-consuming and costly. The identification of molecular markers linked to desirable rootstock traits must first be identified. For example, DNA fragments (genes), may be related to the production of an enzyme that promotes fruit set. If the location of this genetic material (marker) has been determined and the benefits of the marker have been identified, DNA from other rootstocks can be quickly screened for the gene of interest [5]. Parental genetic maps and delineated genomic regions associated with graft (in)-compatibility parameters in apricot has been studied by Pina et al. [6]. Therefore, the selection of desirable rootstocks at the nursery seedling stages can help to reduce the testing duration and exposure required for expensive field trials. Small interfering RNAs (silencing RNA) are now being used to grow virus-resistant transgenic rootstocks [7]. In rootstocks, the potential movement of RNA and DNA genetic or epigenetic factors and the transformation of proteins can be evaluated because of their unique and identifiable characteristics. It is commonly believed that grafted rootstocks and scions maintain their genetic identity, transcription factors, regulatory small interfering RNAs (siRNAs), micro RNAs (miRNAs), mRNAs, peptides, and proteins are mobile in the plant vascular system and may cross the graft union [8,9][8][9].
2. Molecular Mechanism between Scion and Rootstock
Earlier, researchers focused on the role of hormones in vascular reconnection [10]. Now, a successful graft union formation is the function of molecular signaling via phloem which leads to the anatomical and physiological changes in both components for the smooth connectivity of vascular tissues [11,12][11][12]. It is an established fact that the transfer of the intact plastid genome is essential across the graft junction at the molecular level for the uninterrupted communication between the scion and rootstock of the grafted plant [13,14][13][14]. The protein migrates from the companion cells of the shoot into the root cells and controls various physiological processes in plants [9]. The rapid development of molecular biotechnology and “omics” approaches will allow researchers to unravel the physiological and molecular mechanisms involved in the rootstock–scion interaction [15].DNA, RNA, and Protein Transfer during Rootstock–Scion Interaction
As essential components of the RdDM pathway in Arabidopsis, sRNAs migrate bidirectional (from shoot to root and vice versa) [7]. The graft union allows molecules such as DNA, RNA, proteins, and hormones to be transferred bidirectionally between both the component of grafted plants [16,17][16][17]as shown in Figure 1.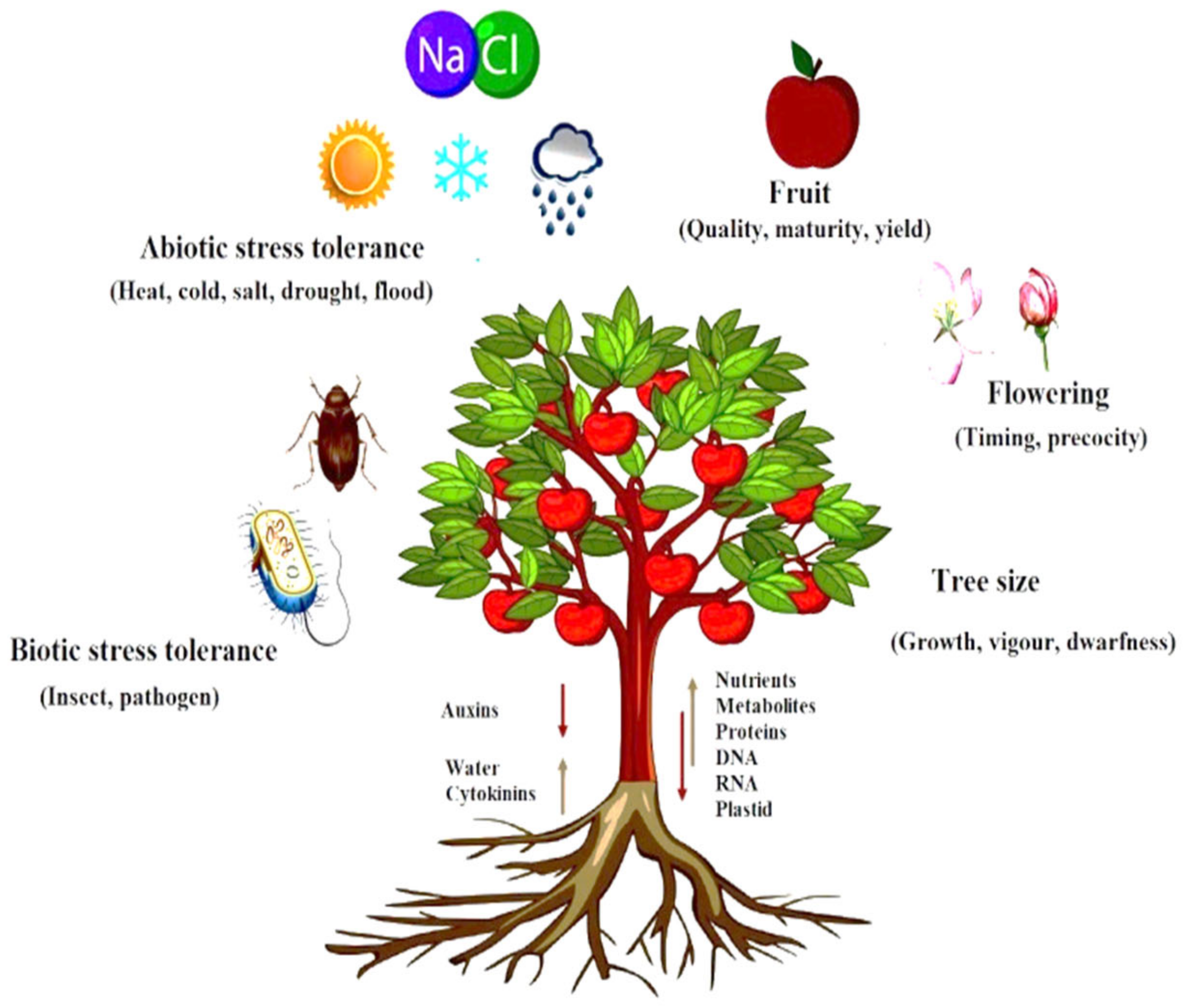
Figure 1. Macromolecules DNA, RNA, protein, hormones, and plastid DNA bidirectional movement via graft union explain the role of molecular and hormone signaling and also explain the effect of scion–rootstock relationship on quality fruit production.
Table 1.
Details of various proteins and their differential expression in horticultural crops.
| Protein | Response | Crop | Reference |
|---|---|---|---|
| CmPP16 | Favoring the process of molecular transport and reduce the degradation of mRNAs. | Cucurbita maxima | [28] |
| PbPTB3 | Play role in long-distance movement of mRNAs across the graft junction by binding of PbPTB3 to PbWoxT1 mRNA. |
Pyrus betlaefolia | [31] |
| Cyclophilin, SICyp1 | Play role in increased auxin response and promoting the growth of roots. | Tomato | [40] |
| PIP1B | Enhanced water levels and cell elongation, leading to better callus formation and successful grafting. | Carya cathayensis | [35] |
| DEPs | At graft unions, 341 and 369 DEPs were found to be upregulated. | Carya cathayensis | [41] |
| PIN | Reunion of vascular tissues is favored by the auxin movement from top to downward direction mediated by PIN proteins. | Arabidopsis | [42] |
References
- Baron, D.; Amaro, A.C.E.; Pina, A.; Ferreira, G. An overview of grafting re-establishment in woody fruit species. Sci. Hortic. 2019, 243, 84–91.
- Faria-Silva, L.; Gallon, C.Z.; Silva, D.M. Photosynthetic performance is determined by scion/rootstock combination in mango seedling propagation. Sci. Hortic. 2020, 265, 109247.
- Dubey, A.K.; Sharma, R.M. Effect of rootstocks on tree growth, yield, quality, and leaf mineral composition of lemon (Citrus limon (L.) Burm.). Sci. Hortic. 2016, 200, 131–136.
- Wang, J.; Jiang, L.; Wu, R. Plant grafting: How genetic exchange promotes vascular reconnection. New Phytol. 2017, 214, 56–65.
- Atkinson, C.; Else, M. Understanding how rootstocks dwarf fruit trees. Compact Fruit Tree 2001, 34, 46–49.
- Pina, A.; Irisarri, P.; Errea, P.; Zhebentyayeva, T. Mapping quantitative trait loci associated with graft (in) compatibility in apriot (Prunus armeniaca L.). Front. Plant Sci. 2021, 12, 622906.
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220.
- Gaffney, T.; Friedrich, L.; Vernooij, B.; Negrotto, D.; Nye, G.; Uknes, S.; Ward, E.; Kessmann, H.; Ryals, J. Requirement of salicylic acid for the induction of systemic acquired resistance. Science 1993, 261, 754–756.
- Paultre, D.S.G.; Gustin, M.P.; Molnar, A.; Oparka, K.J. Lost in transit: Long-distance tracking and phloem unloading of protein signals in Arabidopsishomografts. Plant Cell. 2016, 28, 2016–2025.
- Wetmore, R.H.; Rier, J.P. Experimental induction of vascular tissues in callus of angiosperms. Am. J. Bot. 1963, 50, 418–430.
- Miyashima, S.; Koi, S.; Hashimoto, T.; Nakajima, K. Non-cellautonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 2011, 138, 2303–2313.
- Melnyk, C.W.; Molnar, A.; Bassett, A.; Baulcombe, D.C. Mobile 24nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr. Bio. 2011, 21, 1678–1683.
- Stegemann, S.; Bock, R. Exchange of genetic material between cells in plant tissue grafts. Science 2009, 324, 649–651.
- Stegemann, S.; Keuthe, M.; Greiner, S.; Bock, R. Horizontal transfer of chloroplast genomes between plant species. Proc. Natl. Acad. Sci. USA 2012, 109, 2434–2438.
- Lu, X.; Liu, W.; Wang, T.; Zhang, J.; Li, X.; Zhang, W. Systemic Long-Distance Signaling and Communication between Rootstock and Scion in Grafted Vegetables. Front. Plant Sci. 2020, 11, 460.
- Rasool, A.; Mansoor, S.; Bhat, K.M.; Hassan, G.I.; Baba, T.R.; Alyemeni, M.N.; Ahmad, P. Mechanisms underlying graft union formation and rootstock scion interaction in horticultural plants. Front. Plant Sci. 2020, 11, 1778.
- Kapazoglou, A.; Tani, E.; Avramidou, E.V.; Abraham, E.M.; Gerakari, M.; Megariti, S.; Doupis, G.; Doulis, A.G. Epigenetic changes and transcriptional reprogramming upon woody plant grafting for crop sustainability in a changing environment. Front. Plant Sci. 2021, 11, 613004.
- Taller, J.; Hirata, Y.; Yagishita, N.; Kita, M.; Ogata, S. Graft- induced genetic changes and the inheritance of several characteristics in pepper (Capsicum annuum L.). Theor. Appl. Genet. 1998, 97, 705–713.
- Zhao, L.; Liu, A.; Song, T.; Jin, Y.; Xu, X.; Gao, Y.; Qi, H. Transcriptome analysis reveals the effects of grafting on sugar and α-linolenic acid metabolisms in fruits of cucumber with two different rootstocks. Plant Physiol. Biochem. 2018, 130, 289–302.
- Sharma, A.; Zheng, B. Molecular responses during plant grafting and its regulation by auxins, cytokinins, and gibberellins. Biomoleculess 2019, 9, 397.
- Fuentes, I.; Stegemann, S.; Golczyk, H.; Karcher, D.; Bock, R. Horizontal genome transfer as an asexual path to the formation of new species. Nature 2014, 511, 232–235.
- Molnar, A.; Melnyk, C.W.; Bassett, A.; Hardcastle, T.J.; Dunn, R.; Baulcombe, D.C. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 2010, 328, 872–875.
- Chitwood, D.H.; Timmermans, M.C. Small RNAs are on the move. Nature 2010, 467, 415.
- Sima, X.; Jiang, B.; Fang, J.; He, Y.; Fang, Z.; Km, S.K.; Ren, W.; Qiu, L.; Chen, X.; Zheng, B. Identification by deep sequencing and profiling of conserved and novel hickory microRNAs involved in the graft process. Plant Biotechnol. Rep. 2015, 9, 115–124.
- Kanehira, A.; Yamada, K.; Iwaya, T.; Tsuwamoto, R.; Kasai, A.; Nakazono, M.; Harada, T. Apple phloem cells contain some mRNAs transported over long distances. Tree Genet. Genomes 2010, 6, 635–642.
- Yang, Y.; Mao, L.; Jittayasothorn, Y.; Kang, Y.; Jiao, C.; Fei, Z.; Zhong, G.Y. Messenger RNA exchange between scions and rootstocks in grafted grapevines. BMC Plant Biol. 2015, 15, 251.
- Zhang, Z.; Zheng, Y.; Ham, B.K.; Chen, J.; Yoshida, A.; Kochian, L.V.; Fei, Z.; Lucas, W.J. Vascular-mediated signalling involved in early phosphate stress response in plants. Nat. Plants 2016, 2, 16033.
- Xoconostle-Cazares, B.; Xiang, Y.; Ruiz-Medrano, R.; Wang, H.L.; Monzer, J.; Yoo, B.C.; McFarland, K.C.; Franceschi, V.R.; Lucas, W.J. Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 1999, 283, 94–98.
- Wu, Y.; Cosgrove, D.J. Adaptation of roots to low water potentials by changes in cell wall extensibility and cell wall proteins. J. Exp. Bot. 2000, 51, 1543–1553.
- Lee, Y.; Choi, D.; Kende, H. Expansins: Ever-expanding numbers and functions. Curr. Opin. Plant Biol. 2001, 4, 527–532.
- Duan, X.; Zhang, W.; Huang, J.; Zhao, L.; Ma, C.; Hao, L.; Yuan, H.; Harada, T.; Li, T. KNOTTED1 mRNA undergoes long-distance transport and interacts with movement protein binding protein 2C in pear (Pyrus betulaefolia). Plant Cell Tissue Organ Cult. 2015, 121, 109–119.
- Malz, S.; Sauter, M. Expression of two PIP genes in rapidly growing internodes of rice is not primarily controlled by meristem activity or cell expansion. Plant Mol. Biol. 1999, 40, 985–995.
- Javot, H.; Maurel, C. The role of aquaporins in root water uptake. Ann. Bot. 2002, 90, 301–313.
- Zheng, B.; Liu, L.; Huang, J.; Cheng, X.; Zhu, Y.; Xu, H. Analysis on physiological and biochemical traits of survival of Carya cathayensis grafted seedling. J. Fujian Coll. For. 2002, 22, 320–324.
- Zheng, B.S.; Chu, H.L.; Jin, S.H.; Huang, Y.J.; Wang, Z.J.; Chen, M.; Huang, J.Q. cDNA-AFLP analysis of gene expression in hickory (Carya cathayensis) during graft process. Tree Physiol. 2010, 30, 297–303.
- Xu, D.; Yuan, H.; Tong, Y.; Zhao, L.; Qiu, L.; Guo, W.; Shen, C.; Liu, H.; Yan, D.; Zheng, B. Comparative proteomic analysis of the graft unions in hickory (Carya cathayensis) provides insights into response mechanisms to grafting process. Front. Plant Sci. 2017, 8, 676.
- Kumar, R.S.; Gao, L.X.; Yuan, H.W.; Xu, D.B.; Liang, Z.; Tao, S.C.; Edqvist, J. Auxin enhances grafting success in Carya cathayensis (Chinese hickory). Planta 2018, 247, 761–772.
- Guo, S.; Sun, H.; Tian, J.; Zhang, G.; Gong, G.; Ren, Y.; Zhang, J.; Li, M.; Zhang, H.; Li, H.; et al. Grafting delays watermelon fruit ripening by altering gene expression of ABA centric phytohormone signaling. Front. Plant Sci. 2021, 12, 624319.
- Loupit, G.; Cookson, S.J. Identifying Molecular Markers of Successful Graft Union Formation and Compatibility. Front. Plant Sci. 2020, 11, 610352.
- Spiegelman, Z.; Ham, B.K.; Zhang, Z.; Toal, T.W.; Brady, S.M.; Zheng, Y.; Fei, Z.; Lucas, W.J.; Wolf, S.A. Tomato phloem-mobile protein regulates the shoot-to-root ratio by mediating the auxin response in distant organs. Plant J. 2015, 83, 853–863.
- Yuan, H.; Zhao, L.; Chen, J.; Yang, Y.; Xu, D.; Tao, S.; Zheng, B. Identification and expression profiling of the Aux/IAA gene family in Chinese hickory (Carya cathayensis Sarg.) during the grafting process. Plant Physiol. Biochem. 2018, 127, 55–63.
- Wang, J.; Jin, Z.; Yin, H.; Yan, B.; Ren, Z.; Xu, J.; Mu, C.; Zhang, Y.; Wang, M.; Liu, H. Auxin redistribution and shifts in PIN gene expression during Arabidopsis grafting. Russ. J. Plant Physiol. 2014, 61, 688–696.
More
