Epstein-Barr virus (EBV) was the first human tumor virus to be discovered and is a causative agent for several cancer types of epithelial and lymphoid origin. EBV has two life cycles comprised of latent and lytic phases. The lytic cycle is when new virions are produced, whereas the latent cycle is a state of persistent infection without productive viral replication. It has been recognized that latent infection is the predominant mode of infection in EBV-associated cancers and the expression of a restricted set of latent genes drives disease development. However, we now know that several lytic genes are also expressed in EBV tumors, suggesting a critical role for these genes in tumorigenesis. Here, we summarize the current evidence as to how EBV lytic genes might contribute to EBV-driven oncogenesis.
1. Introduction
EBV is the most prevalent virus infection in humans with approximately 95% of the world’s population sustaining an asymptomatic life-long persistent infection. The discovery that EBV is the causative agent of infectious mononucleosis (IM) represented a breakthrough in our understanding of primary EBV infection in healthy individuals
[1]. A wealth of data demonstrates that EBV is transmitted through saliva with subsequent infection of naïve B cells in the naso/oropharynx. EBV induces proliferation of these B cells into activated B blasts that escape primary T-cell responses and undergo a germinal centre reaction driven through a series of viral latency programs. This culminates in EBV residing in the resting memory B cell pool, thereby establishing a persistent latent infection
[2][3][2,3]. The EBV lytic cycle is switched on when EBV-infected memory B cells differentiate into plasma cells or when the virus enters differentiating epithelial cells, and this results in the production of new virions for transmission to other hosts
[4].
Disruption of this intimate host-viral interaction can lead to malignant transformation. Unlike the pathogenic process in EBV-induced lymphomagenesis where the virus appears to be the initiator, EBV contributes to the development of epithelial tumors, namely nasopharyngeal carcinoma (NPC) and EBV-associated gastric carcinoma (EBVaGC), as a consequence of the aberrant establishment of virus latency in epithelial cells with existing pre-malignant changes that disable the normal differentiation process
[5].
The EBV genome comprises approximately 172 kb and encodes around 100 open reading frames that are generally divided into two categories, latent and lytic genes. A limited subset of viral gene products, the so-called latent proteins, consists of six nuclear antigens (EBNAs 1, 2, 3A, 3B, 3C, and -LP) and three latent membrane proteins (LMPs 1, 2A, and 2B). Over the years, these latent proteins have been regarded as key molecules in the pathogenesis of EBV-associated cancers by regulating various critical cellular processes and pathways
[5]. The contribution of these latent proteins to the oncogenic process remains the subject of intense study. By contrast, although there are more than 80 EBV lytic genes, for the majority of these genes far less attention has been paid to their possible roles in oncogenesis. Nonetheless, it is now becoming increasingly clear that the EBV lytic phase also plays an important role in EBV-driven carcinogenesis. In particular, an early lytic gene, BARF1, is now recognized as being consistently expressed in NPC and EBVaGC, highlighting the need to be more open-minded about the possible contribution of EBV genes outside of those traditionally associated with latent infection. This also stresses the need to obtain more comprehensive EBV gene expression profiles in EBV-associated cancers. The advent of next generation sequencing technology has allowed the discovery and interrogation of exogenous pathogens associated with various types of cancers. RNA sequencing (RNAseq) captures genetic and transcriptional profiles of cancer cells, which can be leveraged to examine not only the host genome but also pathogen genomes infecting host cells. In recent years, an increasing number of research groups have used RNAseq analysis to determine the transcriptomic profiles of EBV-associated cancers
[6][7][8][9][10][11][12][13][14][15][16][17][18][19][6,7,8,9,10,11,12,13,14,15,16,17,18,19]. Although the pattern of EBV gene expression varies between malignancies, these studies consistently demonstrated that a number of EBV lytic genes are more widely expressed than previously recognized, and the expression levels of some of these genes are similar to those of latent genes.
2. EBV Lytic Cycle
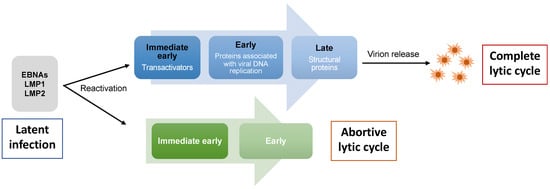
As with many viruses that can establish a persistent infection, the switch from a latent to lytic EBV infection leads to virus replication with the production of new viral particles, and eventually the lysis of productively infected cells. The EBV lytic cycle can be activated in virus-infected B cells and epithelial cells by diverse stimuli in vitro, including phorbol ester and histone deacetylase inhibitors (HDACs)
[20][21][20,21]. While the precise in vivo conditions responsible for this process remain to be explored, it is intimately related to the differentiation of both B cells and epithelial cells. The lytic cycle is characterized by the expression of a large number of viral proteins and is divided into three temporal and functional stages: immediate early, early, and late
[22]. Zta (encoded by
BZLF1) and Rta (encoded by
BRLF1) are the immediate early transcription factors in charge of activating the cascade of EBV lytic gene expression. Early genes mostly encode proteins associated with virus DNA replication, and late gene products mainly support the formation of viral particles (
Figure 1).
Figure 1. EBV lytic cycle. During latent infection, a restricted number of EBV latent genes are expressed in infected cells. Upon reactivation, cells that undergo complete lytic cycle express the immediate early genes (BZLF1, BRLF1), followed by early genes required for viral genome replication. Late genes that encode mainly viral structural proteins are then expressed and this is followed by the production of new viral particles. However, some cells that are reactivated undergo abortive lytic cycle where they express BZLF1 together with one or more early genes in the absence of late genes, and thus do not produce mature virions.
Zta and Rta synergistically stimulate the expression of multiple early lytic genes, including those that encode the components of the core replication machinery: BALF5 (the DNA polymerase), BALF2 (the single-stranded DNA-binding protein homolog), BMRF1 (the DNA polymerase processivity factor), BSLF1 (the primase homolog), BBLF4 (the helicase homolog) and BBLF2/3 (a potential homolog of the third component of the helicase-primase complex)
[23]. In addition to its function as a transactivator, Zta binds to oriLyt (lytic origin of DNA replication) to initiate EBV DNA replication
[24].
Traditionally, it has been thought that the latent and lytic cycles are two mutually exclusive mechanisms that contribute to lifelong infection by EBV. The expression of a restricted set of latent genes is compatible with persistent infection because it allows the virus to escape from host immunity. By contrast, the expression of lytic genes during viral replication is primarily found in the epithelium of oropharynx and salivary glands and it is thought to promote the spread of new viral particles
[25][26][25,26]. It also likely that full lytic infection can occur periodically in B cells, but this has not been demonstrated in vivo. There is a significant immune response, both humoral and cell-mediated, to EBV lytic antigens in all individuals who are infected with EBV. This demonstrates that lytic infection is a continuous feature of virus infection, providing a constant stimulus to the immune system.
A possible tumorigenic role for lytic genes has emerged since the concept of “abortive lytic cycle” was proposed
[27]. Zta preferentially binds and transactivates methylated promoters
[28]. While a complete lytic cycle (i.e., production of progeny virus) immediately post-infection cannot be achieved while the viral genome is not methylated, Zta is still able to induce the expression of several early genes. The expression of
BZLF1 together with one or more early genes in the absence of lytic genes primarily encoding late structural proteins is termed the abortive lytic cycle (
Figure 1).
3. Expression of EBV Lytic Genes in Tumors
The co-existence of latency and abortive lytic cycles has been documented in EBV-associated malignancies, including NPC, EBVaGC, and BL
[29][30][31][29,30,31]. These studies consistently reported the presence of
BZLF1/
BRLF1 and early genes, while late genes were either detected at low frequency or low levels. Unexpectedly, despite evidence for an incomplete lytic cycle, the expression of some late lytic genes has been detected in tumor samples by RNAseq analyses
[6][7][8][9][10][11][12][13][14][15][16][17][18][19][6,7,8,9,10,11,12,13,14,15,16,17,18,19]. Some late genes have been classified as “leaky” because they are expressed at low levels early in the lytic phase and further increased following EBV DNA replication
[32]. However, this does not explain the presence of some “true late genes” in tumor samples whose expression is usually dependent upon lytic DNA replication. Whether EBV acquires a yet-to-be-identified strategy to express late genes that could contribute to the development of disease remains to be determined.
Although there have been reports demonstrating the detection of lytic proteins in a variety of EBV tumors, comprehensive characterization of EBV transcriptomic profiles has only been made possible with the advent of RNAseq analysis. Due to technical and/or clinical challenges, most previous studies have used bulk tumor tissues. Thus, it remains to be determined whether such virus lytic RNA expression was derived from EBV-positive tumor cells or surrounding EBV-positive B cells that happen to be undergoing lytic cycle. Nonetheless, given that several independent research groups reported similar observations across different cancer types, it is now clear that EBV-associated tumors often express a wide-range of lytic genes that could play pivotal roles in EBV oncogenesis. This notion is reinforced by single-cell transcriptomic analysis showing co-existence of EBV latent and lytic genes in tumor cells isolated from primary NPC tissues
[17]. The EBV lytic genes that have consistently been detected in biopsies from different tumor types using RNAseq are listed in
Table 1. Of these, a number have also been detected at the protein level in tissues or cell lines, or antibodies against them detected in patient specimens (
Table 2). As the tumorigenic roles of the two immediate early genes
BZLF1 and
BRLF1 have been discussed and reviewed previously
[33][34][33,34],
rwe
searchers focused
theiour attention on early and late genes with known mechanisms that are relevant to EBV oncogenesis.
Table 1.
Lytic genes that are detected in primary tissues of EBV-associated cancers by RNAseq.
Table 2.
Detection of proteins or antibodies against EBV lytic genes that are expressed in primary EBV-associated cancers by RNAseq.
| Gene Name |
Samples |
Assays |
References |
| BZLF1 |
Biopsy, FNA, cell line |
IHC, ICC, WB |
[31]][31[35],35[36][37,36,37] | [6][7][8][9][10][6,7,8,9,10] |
| BRLF1 |
IE |
| BRLF1 | Transactivator |
GC, NPC, DLBCL |
Plasma, cell line |
ELISA, WB, IF |
[37][38][39][40][37,38,39,40][7][8][10][11][12][7,8,10,11,12] |
| BORF2 |
E |
Ribonucleotide reductase large subunit |
BL, ENKTCL |
[13 |
| BORF2 |
Plasma | ] | [ | 14][13, |
ELISA |
[38]14] |
| BSLF1 |
E |
| BSLF1 |
Cell line | Primase |
GC, DLBCL |
[7 |
WB |
[41]][15][7,15] |
| BSLF2/BMLF1 |
E |
mRNA export factor ICP27 homolog |
PTCL, DLBCL, AITL |
| BSLF2/BMLF1 |
Plasma, cell line |
Protein array, WB |
[38][42][43][38,42,43] | [6][15][16][6,15,16] |
| BALF1 |
E |
vBcl-2 |
GC, NPC |
[8][11][15][17][8,11,15,17] |
| BALF2 |
Biopsy, plasma |
IHC, ELISA |
[38][44][38,44] |
BALF2 |
E |
Single-stranded DNA-binding protein |
GC, NPC, COAD, BL, DLBCL, ENKTCL |
[7][8][9] |
| BHLF1 |
Cell line |
IF |
[45] | [11][14][15][17][18][7,8,9,11,14,15,17,18] |
| BALF3 |
E |
Terminase large subunit |
| BHRF1 |
Biopsy, plasma | GC, NPC, BL, DLBCL, AITL, ENKTCL |
IHC, WB, protein array |
[38][46][47][38,46,47][11][12][14][15][16][17][18][11,12,14,15,16,17,18] |
| BHLF1 |
E |
Involved in viral DNA synthesis |
AITL, BL, DLBCL, |
| BMRF1 |
Biopsy, serum, saliva, cell line |
IHC, ELISA, WB |
[37][48][49][50][37,48,49,50] | [14][15][16][14,15,16] |
| BHRF1 |
| BALF5 | E |
vBcl-2 |
GC, NPC, BL, DLBCL |
Plasma, cell line |
Protein array, WB |
[38][51][38,51[7][9][10][12][14][15][7,9,10,12,14,15] |
| ] |
BMRF1 |
E |
DNA polymerase processivity factor |
GC, COAD, BL, DLBCL, ENKTCL |
[7][9][13][14][15][7,9,13, |
| 9 |
| ] |
| 9 |
| BARF1 |
Biopsy |
WB |
[52] | 14,15] |
| BALF5 |
E |
DNA polymerase catalytic subunit |
GC, NPC, BL, DLBCL, ENKTCL |
[7][8]8[11][12][14],11[,1215,14][,1518][7,,18] |
| BARF1 |
E |
Soluble decoy for CSF-1 |
GC, ENKTCL |
[8][10][11][13][8,10,11,13] |
| BBLF4 |
E |
Helicase |
NPC, GC |
[7][12][7,12] |
| BILF1 |
E |
gp64, vGPCR |
GC, NPC, BL, DLBCL, ENKTCL, |
| BBLF4 |
Cell line |
WB |
[41] |
| BNLF2a |
Cell line |
WB, IF |
[53][54][53,54] |
| LF3 |
FNA, biopsy |
ICC, IHC |
[31][36][31,36] |
[9][11][12][15] |
| BMRF2 |
Cell line |
WB, IF |
[55] | [17][18][9,11,12,15,17,18] |
| BNLF2a |
E |
Inhibtor of TAP |
| BCRF1 |
Allograft, serum | GC, NPC, BL, DLBCL, PTCL, AITL, ENKTCL |
IHC, ELISA |
[56][57][56,57][6][9][11][13][15][16][6[,917,11,13],15,16,17] |
| BNLF2b |
E |
Not reported |
GC, NPC, DLBCL, PTCL, ENKTCL |
[6][11]] |
| BALF4 | [ | 6 | [ | 13, |
Plasma, serum |
Protein array, ELISA |
[38][58][38,11][17][18,13,17,18] |
| 58 | ] |
LF3 |
E |
Involves in viral DNA synthesis |
GC, NPC, BL, AITL |
[11] |
| BKRF2 |
Cell line |
IP |
[59] | [14][16][17][11,14,16,17] |
| BMRF2 |
E/L |
Membrane proteins |
BL, DLBCL, ENKTCL |
| BLLF1 |
Serum, cell line |
IF, WB |
[60][61][62][60,61,62] | [13][14][15][13,14,15] |
| BCRF1 |
L |
vIL-10 |
GC, NPC, AITL, BL, |
[7][9]9[10],10[12],12[16][7,,16] |
| BNRF1 |
Cell line |
WB |
[63] |
BALF4 |
L |
Envelope glycoprotein B |
GC, NPC, BL, DLBCL, ENKTCL |
[7] |
| LF2 | [ | 8 | ] | [9][17][18][7,8,9[11],11[12 |
Plasma],12[14], |
ELISA |
[38]14[15],15,17,18] |
| BKRF2 |
L |
gL, gp25 |
BL, DLBCL |
[14][15][14,15] |
| BLLF1 |
L |
gp350/220 |
GC |
[7][10][7,10] |
| BNRF1 |
L |
Major tegument protein |
GC, NPC, BL, AITL, ENKTCL |
[8][11][12][][18][8,11,1213][14][16,13],14[,1617,17,18] |
| BCLF1 |
L |
Major Capsid Protein |
NPC, GC |
[7][12][7,12] |
| LF1 |
Unknown |
Not reported |
GC, NPC, DLBCL |
[8][11][12][15][17][19][8,11,12,15,17,19] |
| LF2 |
Unknown |
Protein that binds Rta |
GC, NPC, BL, DLBCL |
[8][[11][12][14],11[,1215,14][,1517][8,,17] |