Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Vivi Li and Version 1 by Dhrubajyoti Das.
Seeking optimized infectious pathogen detection tools is of primary importance to lessen the spread of infections, allowing prompt medical attention for the infected. Among nucleic-acid-based sensing techniques, loop-mediated isothermal amplification (LAMP) is a promising method, as it provides rapid, sensitive, and specific detection of microbial and viral pathogens and has enormous potential to transform current point-of-care molecular diagnostics.
- loop-mediated isothermal amplification
- point-of-care
- LAMP-on-a-chip
- pathogen detection
- biosensors
- microfluidic
- digital LAMP
1. Introduction
Point-of-care (POC) testing is performed at the time and location of the healthcare-receiving patients, enabling early-stage diagnosis and prompt medical decisions. Such tests are ideal for environments with limited resources, since they are affordable, need little in the way of equipment, and do away with the requirement for patient follow-up examinations. Nucleic acid amplification tests (NAATs) play an important role in POC diagnosis by delivering pathogen detections at relatively low concentrations, which allows control of infections spreading at the very early stage [1]. In NAATs, the polymerase chain (PCR) reaction is the most commonly used technique and remains a gold standard. However, the dependence on complex thermal cycling, sophisticated instruments, skilled technical personnel, and long reaction time impedes its use in resource-poor settings and for rapid POC testing. Additionally, the advancement of PCR-based approaches for onsite diagnostics is further hampered by non-specific amplification and a higher risk of false positives [1,2][1][2]. Numerous state-of-the-art technologies have been developed in recent years for rapid pathogen detection, such as the enzyme-linked immunosorbent assay (ELISA) [3], surface-enhanced Raman spectroscopy (SERS) [4], surface plasmon resonance (SPR) [5], electrochemical biosensors [6], and clustered regularly interspaced short palindromic repeats (CRISPR) [7]. Although these technologies are promising, they are currently unable to fully address the difficulties associated with pathogen detection, since their implementation in areas with low resources is hampered by the need for extensive pre-processing, sophisticated equipment, and pricey labeling. There is still a demand for developing integrated biosensors that offer rapid and precise detection of infectious pathogens with high sensitivity [8].
Isothermal nucleic acid amplification tests (iNAATs) have been developed as an alternative and alluring method for highly accurate, quick, and economically efficient nucleic acid amplification [9]. Isothermal amplification (IA) works with a constant temperature (35–65 °C) with a reaction time of 60–90 min. Therefore, it only requires simple hardware for heating and is easy to integrate with a microfluidic device. In addition, IA has higher tolerance toward enzyme inhibitors, making this a robust amplification method [10]. This method includes loop-mediated isothermal amplification (LAMP) [11], recombinase polymerase amplification (RPA) [12], rolling circle amplification (RCA) [13], nucleic-acid-sequence-based amplification (NASBA) [14], and helicase-dependent amplification (HDA) [15]. Among them, LAMP is considered the most promising and robust due to the formation of large copy numbers in a short period (20–30 min) from low concentrations of target molecules. In particular, the amount of amplicon produced from a LAMP reaction is >50-fold more than any other PCR-based amplification technique. LAMP also has the potential to amplify multiple sizes of target DNA ranging from 130–300 bp, delivering amplification for versatile DNA of pathogens [11]. The reverse transcription LAMP (RT-LAMP) technique is employed to amplify RNA molecules using a reverse transcriptase enzyme to synthesize cDNA out of RNA. LAMP has additional advantages compared to other IA; for example: (i) RPA requires a recombinase enzyme, a protein for single-stranded DNA binding, and a polymerase enzyme for strand displacement, but in LAMP, a single DNA polymerase enzyme is used [16]; (ii) LAMP is a one-step reaction process, whereas in RCA, an additional ligation step is required before the amplification [17]; (iii) LAMP amplifies both DNA and RNA molecules—however, NASBA is used to amplify only RNA molecules [10]; (iv) unlike other IA methods, LAMP uses 4–6 primers makes it highly specific [18]; and (v) LAMP also exhibits very high tolerance to polymerase inhibitors, making it highly desirable for direct analysis of body fluids and other clinical samples, without further sample enrichments [19,20][19][20]. In addition, the handy design guidelines of LAMP attracted more users, which helped with creating a strong research database over the past two decades. Furthermore, the patent-free status makes LAMP available for the development of commercial products for rapid and onsite diagnosis of infectious diseases at POC.
2. Overview of the LAMP Assay
2.1. Principle
Isothermal amplification (IA) is a simple, rapid, and efficient method that can accumulate nucleic acid at a constant temperature without using additional advanced high-tech equipment. Due to the operational simplicity, IA methods are easy to integrate with microfluidic chips to develop rapid, sensitive, and onsite POC diagnostic tools [9]. Among the IA methods developed during 1991–2006, LAMP has risen as a rapid, robust, and highly specific nucleic acid amplification technique with the ability to generate a large number of products within a short time in a single-step reaction. LAMP was first reported in the year 2000 by Notomi et al. [11], and since, it has gained popularity in research, clinical, and industrial applications and emerged as an alternative to conventional PCR by eliminating the dependence on an expensive thermocycler. Additionally, the high tolerance of LAMP towards the enzyme inhibitors makes it a suitable candidate for onsite detection of clinical and biological samples without any sample enrichment. Unlike conventional PCR, LAMP requires four specific primers targeting six distinct regions (F3c, F1c, F2c, B2c, B1c, and B3c) on the DNA, providing enhanced specificity. This set of primers includes two inner primers called forward inner primer (FIP) and backward inner primer (BIP), whereas F3 and B3 are two outer primers, also known as “displacement primers” [23][21]. Figure 1 displays the mechanism of the LAMP reaction [24][22]. Both the inner primers FIP and BIP consist of hybridization sequences, F2 and F1c and B2 and B1c, respectively. The inner primers contain a unique “fold back” arrangement that creates stem–loop motifs for self-priming [11]. LAMP amplification can be split into two steps, i.e., the structure creation and the cyclic amplification. LAMP benefits from a DNA polymerase with strand-displacing activity, which avoids the need for a complex heat cycling step for dsDNA denaturation between each amplification cycle. In the first step, the F2 of the inner primer binds with the F2c of the target stand, initiating DNA synthesis due to the polymerase activity. Next, the F3 of the outer primer binds with F3c, followed by the displacement of the freshly synthesized DNA and the release of the target DNA. The released strand containing complementary F1 and F1c regions gives rise to a stem–loop structure. In the 3′ ends, another stem–loop structure will be formed under a similar mechanism to BIP and B3, forming a “dumbbell”-like structure. This “dumbbell structure” will undergo self-priming and serve as a starting material for the cyclic amplification step. As a result, LAMP produces a significant number of amplicons in a rapid period. Due to these characteristics, LAMP is well-suited for usage in POC biosensors that feature simplicity, resilience, miniature, and ease of operation.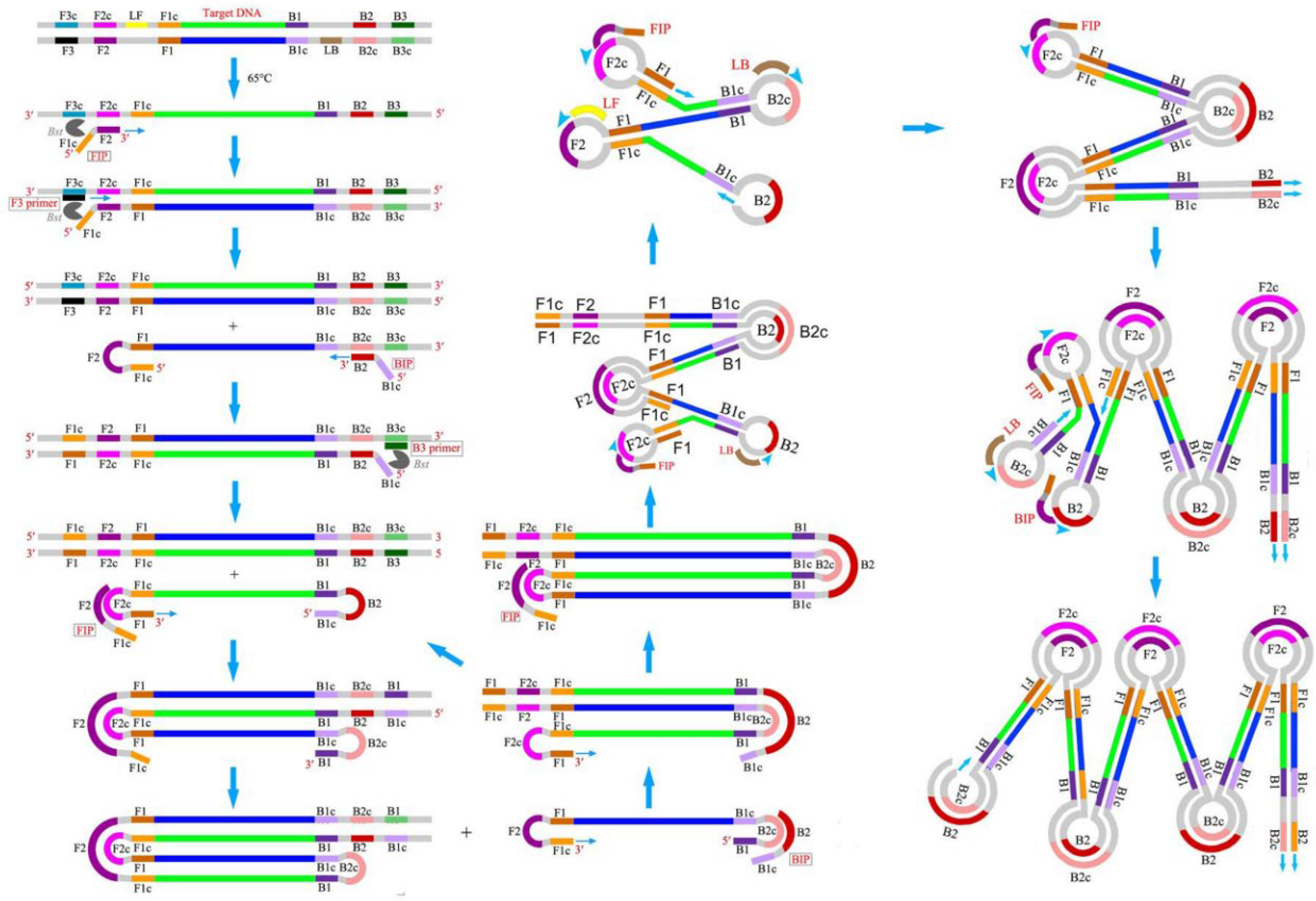
Figure 1. Schematic of LAMP mechanism. Reproduced with permission from reference [24], open access. Copyright 2016 Frontiers Media.
Schematic of LAMP mechanism. Reproduced with permission from reference [22], open access. Copyright 2016 Frontiers Media.
2.2. Primer Design
In addition to the four basic primers (FIP, BIP, F3, and B3), LAMP utilizes two other loop primers, the forward loop primer (LF) and backward loop primer (LB) [18], for the acceleration of the amplification and reduction of the time to one-third of the usual. The loop region is typically situated between B1–B2 and F1–F2 of the dumbbell structure, providing additional initiation sites for LAMP. The basic principles of LAMP primer designs are as follows: (i) there should be a minimum distance of 40–60 bases between F2 and F1c and B2 and B1c for enough space to generate loop primers; (ii) repeats of dinucleotides (e.g., AGAGAG) and repetition of a single nucleotide more than three times (e.g., ATTTT) should be avoided, as it can cause a mispriming; (iii) three or more runs of Gs should be avoided, as it may raise an issue with primer synthesis and purification; (iv) to avoid the secondary structure formation, the 3′ ends of the designed primers should not be AT-rich or complementary to one another; (v) the Tm for the primer pairs (e.g., F2 and B2) should be similar to a maximum temperature difference of 5 °C, and the F1c and B1c should have a higher Tm than F2 and B2 for the immediate formation of the stem–loop structures; (vi) in the case of GC-rich primers, the melting temperature (Tm) should be about 60–65 °C, and in the case of AT-rich primers, it should be around 55–60 °C. A relatively higher Tm is required for GC-rich primers due to the formation of a higher number of hydrogen bonds. The Tm of the primers is calculated using Equation (1); (vii) to obtain higher stability of the primers, the ΔG values of the 5′ ends of F1c/B1c and 3′ ends of F2/B2 and F3/B3, should be less than −4 kcal/mol; and (viii) to eliminate the formation of secondary structures, the 3′ ends should not be AT-rich [25][23]. NUPACK software (version 4.0, created by Prof. Niles A. Pierce and his team, Copyright© 2006–2022 California Institute of Technology, http://www.nupack.org/partition/new, accessed on 20 July 2022) is used to analyse putative secondary structures, and primer dimer production and hybridization stability [26][24]. In addition to that, reports suggest that the integration of poly T linkers (TTTT) between F2 and F1c of FIP and between B2 and B1c of BIP can improve the reaction speed by loop formation [27][25]. where R is the gas constant 1.9872 × 10−3 kcal K−1 mol−1; the number in the second term, −273.15, is a conversion factor from K to °C; CT is the total molar concentration (M) of the strands; and X is considered four for non-self-complementary duplexes.
Primer explorer V5 (http://primerexplorer.jp/elamp4.0.0/index.html, accessed on 20 July 2022) is the most commonly used software for LAMP primer design. The selected target region should be 200–2000 bp in length, and the saved sequence can only be uploaded in FASTA, .txt, or GenBank format. The steps for designing LAMP primers using Primer Explorer V5 are as follows: (i) launch the program, upload the correct format of the sequence file, and click on “Primer Design” to display the regions of the target sequences; (ii) click on “Generate” to get the designed primers of the best five primer options for a particular gene sequence; (iii) choose the primer set with the highest ΔG value for dimerization and click on “Display” to exhibit the designed primer list in a new window; (iv) select the primer with a ΔG value of ≤−4 kcal/mol at the 5′ end of F1c/B1c and at the 3′ end of F2/B2 and F3/B3 and click on “Confirm” to obtain the particular primer details; (v) after choosing the appropriate primer set, click on “Primer information” to save the file for loop-primer generation, and then click on “Save” to download the primer sequences in a Word/Excel document; (vi) to generate the loop primers, re-launch the software and upload the saved file from step (v); (vii) click on “Primer Design” to display the LF and LB in the target sequence; (viii) next, click on “Generate,” followed by “Display,” to obtain the designed LF and LB primers in a new window; (ix) choose the loop primer pair with the highest ΔG value for dimerization and click on “Confirm” to display the primer details; (x) finally, choose the loop primers with lowest ΔG value at 3′ end and click on “Save” to download the primers. BLAST tool (www.ncbi.nlm.nih.gov, accessed on 20 July 2022) is used to check the specificity of each of the designed primers. The designed primer should display high specificity for the targeted gene sequence. The software also provides advanced settings with the flexibility of choosing AT and GC-rich options and the desired Tm to design target-specific customized primers. Nevertheless, it is not advised to change the lengths of primers. Primer explorer V5 is an easy-to-use online tool for LAMP primer design; however, it has some limitations—e.g., (i) simultaneous design of the loop primer is not possible; (ii) it only allows a gene sequence up to 2000 bp; (iii) besides ATGC, it does not support any other IUPAX characters; and (iv) it allows only one execution process.
Apart from the primer explorer, multiple online primer design software tools are available, such as Premier Biosoft, Optigene LAMP designer, NEB LAMP primer designer tool, and more. Several methods have been published in recent years for designing target-based primers. For example, Zhang et al. [28][26] developed a novel Python script for designing LAMP-based variant-specific probes to detect cancer mutations, evaluated using the sequences of ESR1 p.E380Q and ESR1 p.Y537S cancer-specific mutations, as the algorithm generates the two best primer sets for the respective target with an input of the DNA sequence. In another work, Jia et al. [29][27] presented a whole genome-based LAMP primer design (GLAPD) tool, which can be downloaded using the following website: http://cgm.sjtu.edu.cn/GLAPD/ or https://github.com/jiqingxiaoxi/GLAPD.git (accessed on 20 July 2022) and http://cgm.sjtu.edu.cn/GLAPD/online/ (accessed on 20 July 2022). It is a novel approach that uses complete genomes to create LAMP primers for a set of target genomes for rapid detection of foodborne pathogens, which yielded similar results to Primer explorer V5 with the same nucleotide sequences. Additionally, a theoretical approach was developed by Savonnet et al. [30][28], who studied the dynamics of LAMP’s second stage from the fundamentals of reaction rates to deduce physio-chemical properties from the experimental signal development. The dumbbell structure used in the study skipped the first stage of the LAMP. This model anticipates that the concentration of amplification products will rise according to a logistic function.
where R is the gas constant 1.9872 × 10−3 kcal K−1 mol−1; the number in the second term, −273.15, is a conversion factor from K to °C; CT is the total molar concentration (M) of the strands; and X is considered four for non-self-complementary duplexes.
Primer explorer V5 (http://primerexplorer.jp/elamp4.0.0/index.html, accessed on 20 July 2022) is the most commonly used software for LAMP primer design. The selected target region should be 200–2000 bp in length, and the saved sequence can only be uploaded in FASTA, .txt, or GenBank format. The steps for designing LAMP primers using Primer Explorer V5 are as follows: (i) launch the program, upload the correct format of the sequence file, and click on “Primer Design” to display the regions of the target sequences; (ii) click on “Generate” to get the designed primers of the best five primer options for a particular gene sequence; (iii) choose the primer set with the highest ΔG value for dimerization and click on “Display” to exhibit the designed primer list in a new window; (iv) select the primer with a ΔG value of ≤−4 kcal/mol at the 5′ end of F1c/B1c and at the 3′ end of F2/B2 and F3/B3 and click on “Confirm” to obtain the particular primer details; (v) after choosing the appropriate primer set, click on “Primer information” to save the file for loop-primer generation, and then click on “Save” to download the primer sequences in a Word/Excel document; (vi) to generate the loop primers, re-launch the software and upload the saved file from step (v); (vii) click on “Primer Design” to display the LF and LB in the target sequence; (viii) next, click on “Generate,” followed by “Display,” to obtain the designed LF and LB primers in a new window; (ix) choose the loop primer pair with the highest ΔG value for dimerization and click on “Confirm” to display the primer details; (x) finally, choose the loop primers with lowest ΔG value at 3′ end and click on “Save” to download the primers. BLAST tool (www.ncbi.nlm.nih.gov, accessed on 20 July 2022) is used to check the specificity of each of the designed primers. The designed primer should display high specificity for the targeted gene sequence. The software also provides advanced settings with the flexibility of choosing AT and GC-rich options and the desired Tm to design target-specific customized primers. Nevertheless, it is not advised to change the lengths of primers. Primer explorer V5 is an easy-to-use online tool for LAMP primer design; however, it has some limitations—e.g., (i) simultaneous design of the loop primer is not possible; (ii) it only allows a gene sequence up to 2000 bp; (iii) besides ATGC, it does not support any other IUPAX characters; and (iv) it allows only one execution process.
Apart from the primer explorer, multiple online primer design software tools are available, such as Premier Biosoft, Optigene LAMP designer, NEB LAMP primer designer tool, and more. Several methods have been published in recent years for designing target-based primers. For example, Zhang et al. [28][26] developed a novel Python script for designing LAMP-based variant-specific probes to detect cancer mutations, evaluated using the sequences of ESR1 p.E380Q and ESR1 p.Y537S cancer-specific mutations, as the algorithm generates the two best primer sets for the respective target with an input of the DNA sequence. In another work, Jia et al. [29][27] presented a whole genome-based LAMP primer design (GLAPD) tool, which can be downloaded using the following website: http://cgm.sjtu.edu.cn/GLAPD/ or https://github.com/jiqingxiaoxi/GLAPD.git (accessed on 20 July 2022) and http://cgm.sjtu.edu.cn/GLAPD/online/ (accessed on 20 July 2022). It is a novel approach that uses complete genomes to create LAMP primers for a set of target genomes for rapid detection of foodborne pathogens, which yielded similar results to Primer explorer V5 with the same nucleotide sequences. Additionally, a theoretical approach was developed by Savonnet et al. [30][28], who studied the dynamics of LAMP’s second stage from the fundamentals of reaction rates to deduce physio-chemical properties from the experimental signal development. The dumbbell structure used in the study skipped the first stage of the LAMP. This model anticipates that the concentration of amplification products will rise according to a logistic function.
2.3. Features for Rapid Pathogen Detection
Due to the formation of high copy numbers (~109) in a short period (<1 h), LAMP has been considered one of the most efficient and popular diagnostics tools used for rapid pathogen detection. The inner primer plays a crucial role in LAMP, as it has brought a lot of benefits for making LAMP a robust, rapid, and advanced amplification method. Essentially, the strand displacement of the inner primer occurs at a constant temperature (60–65 °C), eliminating the need for complex thermal cycling and making LAMP a suitable candidate for POC devices with its simple design and rapidness. The stem–loop structure provides amplification based on self-priming, allowing the production of large numbers of amplicons independent of template DNA strands. Hence, this produces a high copy number of DNA in a short period, enabling rapid target detection with very high sensitivity. LAMP provides a high tolerance towards the inhibitors, reducing the effort of sample pre-treatment. Therefore, the idea of a simple LAMP-based “sample-in-result-out” POC device is feasible. Compared to other methods, LAMP uses 4–6 different primers targeting six distinctive regions, making it highly specific. A mismatch with any target sequences will lead to a negative amplification, eliminating the chances of non-specific amplification. It is reported that the LAMP assay may possess higher sensitivity compared to the gold-standard PCR. It can have a detection limit as low as five copies per reaction [31][29]. Hence, LAMP can be used for early disease diagnosis and to contain the spread of infectious pathogens. Furthermore, the production of pyrophosphate molecules during LAMP enables visual identification of the target, making the detection method easy and simple without using any expensive instrumentation. As another factor, LAMP produces polymerized target DNA with various base pair lengths from 300 to 25 kb [11]. This large number of amplified nucleic acids could increase the viscosity of solutions. This micro-level viscosity change can be detected by an advanced and ultrasensitive particle-imaging technique called rotational diffusometry, bringing down the detection time to less than 10 min [32][30]. A detailed discussion is included in Section 3.1.5. Another fascinating tool named CRISPR/Cas system [33][31] has been integrated with LAMP for rapid detection of pathogenic nucleic acids (NA) with very high sensitivity [34][32]. First, the Cas protein (Cas9, Cas12, and Cas13) forms a complex with a special RNA sequence called CRISPR-RNA (crRNA), activated once it binds to the specific target RNA/DNA, triggering the cleaving mechanism of Cas protein. Subsequently, this activated complex cleaves the reporter probe carrying a fluorophore and a quencher, releasing significant fluorescence. The detection can be performed via monitoring fluorescence and in lateral-flow strips [35,36][33][34]. Recently, CRISPR, in combination with RT-LAMP, has been investigated for rapid and highly accurate POC diagnosis of COVID-19 [37,38][35][36].3. Application of LAMP in Clinical Diagnosis
Access to high-quality medical care and technologies is still a major issue in resource-constrained places, particularly in low- and middle-income countries. Therefore, it is crucial to develop POC devices that offer an affordable, trustworthy, and quick screening of medical diseases outside of the laboratory setting. The World Health Organization (WHO) has established standards for assessing POC diagnostic platforms, with an acronym “ASSURED”—meaning they should have the following qualities: being low-cost/affordable, highly sensitive and specific (very low false positive), user-friendly (anyone can use), rapid/robust, high-tech equipment-free, and the deliverable (portable). Due to its operational simplicity, LAMP is one of the best candidates to fulfil these criteria. Therefore, LAMP has been massively used in molecular diagnostics for various infectious diseases, especially during the time of COVID-19. For example, Song et al. [160][37] developed an instrument-free, two-stage isothermal amplification method called Penn-RAMP, combining reverse-transcription recombinase polymerase amplification (RT-RPA) and LAMP for the detection of SARS-CoV-2, targeting ORF1ab gene with improved efficiency. In this method, the RT-RPA is performed first in the cap of a reaction tube. The reaction was carried out at 38 °C with nasopharyngeal swabs and saliva samples. F3 and B3 LAMP primers and a recombinase polymerase enzyme were used for the RPA reaction. After 15–20 min, the RPA reaction mixture was transferred to the pre-loaded LAMP mixture in the same reaction tube. In the second stage, the LAMP reaction was performed at 63 °C for 40 min. RPA and LAMP mixtures were maintained in a 1:9 ratio to prevent LAMP inhibition due to RPA contents. The assay provided a LOD of 5 copies/reaction with a 10-fold higher sensitivity than RT-LAMP. Hence, Penn-RAMP overcomes the individual limitations of RPA and LAMP and delivers higher sensitivity. Additionally, integration of the CRISPR/Cas diagnostic method with LAMP provides the detection of pathogenic nucleic acids with high sensitivity and specificity. For example, Joung et al. [161][38] have developed STOPCovid (SHERLOCK testing in one-pot for COVID-19), a CRISPR/Cas-based one-pot detection method of SARS-CoV-2 combined with RT-LAMP. In this method, first, the viral RNA is isothermally amplified; then the amplicons are cleaved by a CRISPR-mediated reporter and sequentially detected. Viral RNA is extracted by thermal lysis at 60 °C for 10 min in a commercial lysis buffer. Thus, this method eliminates the additional viral RNA extraction step. After a 60 min reaction, the final product is identified by lateral flow strip or fluorescent reporter. The assay achieved a LOD of 100 copies/reaction in just 70 min. It also delivered very high accuracy (11/12) for clinical samples. Similarly, Broughton et al. [162][39] reported a rapid RT-LAMP-based SARS-CoV-2 detection platform named DETECTR (SARS-CoV-2 DNA Endonuclease-Targeted CRISPR Trans Reporter) with integrated CRISPR/Cas12 detection. This assay performs the RT-LAMP targeting of N (nucleoprotein) and E (envelop) genes of SARS-CoV-2, followed by Cas12 recognition of predetermined viral RNA sequences and virus detection on a lateral flow strip upon cleavage of the reporter molecule. A LOD of 10 copies/µL was achieved in just 40 min. Clinical samples were tested by extracting RNA from oropharyngeal/nasopharyngeal swabs of COVID-19 patients. The DETECTR assay showed 95% accuracy relative to the gold standard, RT-PCR.FDA-Approved LAMP-Based Devices
Various LAMP-based commercial POC test kits for SARS-CoV-2 were developed and received emergency use authorization (EUA) from the U.S. Food and Drug Administration (FDA). The Lucira COVID-19 All-in-one Test Kit was developed by Lucira Health Inc. [163][40] and received the EUA on 17 November 2020 for prescription domestic COVID-19 testing [164][41], and on 9 April 2021, the Lucira Check-it COVID-19 Test received authorization by the FDA for non-prescription home use [165][42]. It utilizes the RT-LAMP by targeting the N-gene of SARS-CoV-2 RNA. The colorimetric detection based on the pH change of the sample due to LAMP delivers the detection within 30 min. The nasal swab is dissolved in the elution buffer, which provides the viral lysis at room temperature. The buffer solution dissolves the preloaded lyophilized reagents, and the colorimetric readout is delivered by an electronic and optical element in the test kit. No pumps are needed because the device uses capillary flow and gravity, and it just has one user-activated valve. The heater and optics are located on a single PCB. A LOD of 2700 copies/swab was achieved for Lucira Check-It, with a detection accuracy of 98%. Similarly, Detect Inc. combined RT-LAMP with lateral flow technology and developed the Detect COVID-19 test kit [166][43]. This test kit targets the ORF1ab gene of the SARS-CoV-2 viral genome. The Detect Hub is used in conjunction with the disposable test tube and collection buffer. After connecting, the Detect Hub must be kept aside for 65 min before the test can be conducted, likely to stabilize the system. The RT-LAMP reaction will begin automatically when the tube has been inserted into the Detect Hub. The amplification takes about 55 min, followed by a lateral flow assay for 10 min, and the read-out of the result is performed using the Detect app on a smartphone. The kit provides a human positive control to confirm the appropriate sample collection and the viral RNA extraction. Although the Detect COVID-19 test is intended to be used at home, only healthcare professionals could purchase IFU. On 11 April 2022, the FDA issued a letter of authorization to Detect Inc. [167][44]. The test may be used at home without a prescription. Table 51 listed the recently developed LAMP-based molecular diagnostic methods for SARS-CoV-2 detection and provides a comparison between them.Table 51.
RT-LAMP-based methods for COVID-19 diagnosis.
| Method | Target Gene | Time | Sensitivity | Specimen (Swab Type) | LOD | EUA by FDA | Price Per Unit |
|---|---|---|---|---|---|---|---|
| STOP Covid [161][38] | N gene | 70 min | 91.6% | Nasopharyngeal and oropharyngeal swabs | 100 copies/reaction | No | $40 USD |
| Penn-RAMP [160][37] | ORF1ab and N gene | 60 min | 84% | Nasopharyngeal swab and saliva | 5 copies/reaction | No | NA |
| DETECTR [162][39] | N and E gene | 40 min | 95% | Nasopharyngeal and oropharyngeal swabs | 10 copies/µL | No | NA |
| iSCAN [168][45] | N and E gene | 60 min | 86% | Nasopharyngeal swab | 10 copies/sample | No | $2–5 USD |
| iLACO [169][46] | ORF1ab gene | 40 min | 89.9% | -- | 10 copies/µL | No | NA |
| Lucira Check-it [165][42] | N gene | 30 min | 98% | Nasal swab | 2700 copies/swab | Yes | $68 USD |
| Detect COVID-19 test [166][43] | ORF1ab gene | 65 min | 95% | Nasal swab | 800 copies/mL | Yes | $55 USD |
| Metrix COVID-19 test [170][47] | ORF1ab and N gene | 30 min | 95% | Nasal swabs and saliva | 667 copies/mL | Yes | NA |
| DxLab COVID-19 test [171][48] | M gene | 25 min | 95% | Nasal swab | 3000 copies/swab | Yes | NA |
References
- Yang, S.; Rothman, R. PCR-based diagnostics for infectious diseases: Uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 2004, 4, 337–348.
- Demeke, T.; Jenkins, G.R. Influence of DNA extraction methods, PCR inhibitors and quantification methods on real-time PCR assay of biotechnology-derived traits. Anal. Bioanal. Chem. 2009, 396, 1977–1990.
- Zhao, Y.; Zeng, D.; Yan, C.; Chen, W.; Ren, J.; Jiang, Y.; Jiang, L.; Xue, F.; Ji, D.; Tang, F.; et al. Rapid and accurate detection of Escherichia coli O157:H7 in beef using microfluidic wax-printed paper-based ELISA. Analyst 2020, 145, 3106–3115.
- Yang, Y.; Li, G.; Wang, P.; Fan, L.; Shi, Y. Highly sensitive multiplex detection of foodborne pathogens using a SERS immunosensor combined with novel covalent organic frameworks based biologic interference-free Raman tags. Talanta 2022, 243, 123369.
- Lee, N.; Choi, S.-W.; Chang, H.-J.; Chun, H.S. Rapid Detection of Escherichia coli O157:H7 in Fresh Lettuce Based on Localized Surface Plasmon Resonance Combined with Immunomagnetic Separation. J. Food Prot. 2018, 81, 713–718.
- Kaya, H.O.; Cetin, A.E.; Azimzadeh, M.; Topkaya, S.N. Pathogen detection with electrochemical biosensors: Advantages, challenges and future perspectives. J. Electroanal. Chem. 2021, 882, 114989.
- Tian, Y.; Liu, T.; Liu, C.; Xu, Q.; Liu, Q. Pathogen detection strategy based on CRISPR. Microchem. J. 2022, 174, 107036.
- Nesakumar, N.; Lakshmanakumar, M.; Srinivasan, S.; Jbb, A.J.; Rayappan, J.B.B. Principles and Recent Advances in Biosensors for Pathogens Detection. ChemistrySelect 2021, 6, 10063–10091.
- Pumford, E.A.; Lu, J.; Spaczai, I.; Prasetyo, M.E.; Zheng, E.M.; Zhang, H.; Kamei, D.T. Developments in integrating nucleic acid isothermal amplification and detection systems for point-of-care diagnostics. Biosens. Bioelectron. 2020, 170, 112674.
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545.
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63.
- Piepenburg, O.; Williams, C.H.; Stemple, D.; Armes, N. DNA Detection Using Recombination Proteins. PLOS Biol. 2006, 4, e204.
- Liu, D.; Daubendiek, S.L.; Zillman, M.A.; Ryan, K.; Kool, E.T. Rolling Circle DNA Synthesis: Small Circular Oligonucleotides as Efficient Templates for DNA Polymerases. J. Am. Chem. Soc. 1996, 118, 1587–1594.
- Compton, J. Nucleic acid sequence-based amplification. Nature 1991, 350, 91–92.
- Vincent, M.; Xu, Y.; Kong, H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004, 5, 795–800.
- Suea-Ngam, A.; Bezinge, L.; Mateescu, B.; Howes, P.D.; Demello, A.J.; Richards, D.A. Enzyme-Assisted Nucleic Acid Detection for Infectious Disease Diagnostics: Moving toward the Point-of-Care. ACS Sens. 2020, 5, 2701–2723.
- Xu, L.; Duan, J.; Chen, J.; Ding, S.; Cheng, W. Recent advances in rolling circle amplification-based biosensing strategies-A review. Anal. Chim. Acta 2020, 1148, 238187.
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 2002, 16, 223–229.
- Iseki, H.; Kawai, S.; Takahashi, N.; Hirai, M.; Tanabe, K.; Yokoyama, N.; Igarashi, I. Evaluation of a Loop-Mediated Isothermal Amplification Method as a Tool for Diagnosis of Infection by the Zoonotic Simian Malaria Parasite Plasmodium knowlesi. J. Clin. Microbiol. 2010, 48, 2509–2514.
- Abdul-Ghani, R.; Al-Mekhlafi, A.M.; Karanis, P. Loop-mediated isothermal amplification (LAMP) for malarial parasites of humans: Would it come to clinical reality as a point-of-care test? Acta Trop. 2012, 122, 233–240.
- Liu, A.; Guan, G.; Du, P.; Gou, H.; Liu, Z.; Liu, J.; Ma, M.; Yang, J.; Li, Y.; Niu, Q.; et al. Loop-mediated isothermal amplification (LAMP) method based on two species-specific primer sets for the rapid identification of Chinese Babesia bovis and B. bigemina. Parasitol. Int. 2012, 61, 658–663.
- Li, J.-J.; Xiong, C.; Liu, Y.; Liang, J.-S.; Zhou, X.-W. Loop-Mediated Isothermal Amplification (LAMP): Emergence as an Alternative Technology for Herbal Medicine Identification. Front. Plant Sci. 2016, 7, 1956.
- Parida, M.; Sannarangaiah, S.; Dash, P.K.; Rao, P.V.L.; Morita, K. Loop mediated isothermal amplification (LAMP): A new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 2008, 18, 407–421.
- Zadeh, J.N.; Steenberg, C.D.; Bois, J.S.; Wolfe, B.R.; Pierce, M.B.; Khan, A.R.; Dirks, R.M.; Pierce, N.A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 32, 170–173.
- Li, Y.; Fan, P.; Zhou, S.; Zhang, L. Loop-mediated isothermal amplification (LAMP): A novel rapid detection platform for pathogens. Microb. Pathog. 2017, 107, 54–61.
- Zhang, J.; Alexandrou, G.; Toumazou, C.; Kalofonou, M. Automating the Design of Cancer Specific DNA Probes Using Computational Algorithms. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society, Guadalajara, Mexico, 1–5 November 2021; pp. 1852–1856.
- Jia, B.; Li, X.; Liu, W.; Lu, C.; Lu, X.; Ma, L.; Li, Y.-Y.; Wei, C. GLAPD: Whole Genome Based LAMP Primer Design for a Set of Target Genomes. Front. Microbiol. 2019, 10, 2860.
- Savonnet, M.; Aubret, M.; Laurent, P.; Roupioz, Y.; Cubizolles, M.; Buhot, A. Kinetics of Isothermal Dumbbell Exponential Amplification: Effects of Mix Composition on LAMP and Its Derivatives. Biosensors 2022, 12, 346.
- Craw, P.; Balachandran, W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: A critical review. Lab Chip 2012, 12, 2469–2486.
- Das, D.; Chen, W.-L.; Chuang, H.-S. Rapid and Sensitive Pathogen Detection by DNA Amplification Using Janus Particle-Enabled Rotational Diffusometry. Anal. Chem. 2021, 93, 13945–13951.
- Liu, J.-J.; Orlova, N.; Oakes, B.L.; Ma, E.; Spinner, H.B.; Baney, K.L.M.; Chuck, J.; Tan, D.; Knott, G.J.; Harrington, L.B.; et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 2019, 566, 218–223.
- Selvam, K.; Najib, M.A.; Khalid, M.F.; Mohamad, S.; Palaz, F.; Ozsoz, M.; Aziah, I. RT-LAMP CRISPR-Cas12/13-Based SARS-CoV-2 Detection Methods. Diagnostics 2021, 11, 1646.
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442.
- Hatoum-Aslan, A. CRISPR Methods for Nucleic Acid Detection Herald the Future of Molecular Diagnostics. Clin. Chem. 2018, 64, 1681–1683.
- Bhatt, A.; Bumbrah, G.S.; Ruwali, M.; Hameed, S.; Fatima, Z. Diagnostic efficiency of RT-LAMP integrated CRISPR-Cas technique for COVID-19: A systematic review and meta-analysis. Pathog. Glob. Health 2022, 116, 625.
- Zhang, Y.; Chen, M.; Liu, C.; Chen, J.; Luo, X.; Xue, Y.; Liang, Q.; Zhou, L.; Tao, Y.; Li, M.; et al. Sensitive and rapid on-site detection of SARS-CoV-2 using a gold nanoparticle-based high-throughput platform coupled with CRISPR/Cas12-assisted RT-LAMP. Sens. Actuators B Chem. 2021, 345, 130411.
- Song, J.; El-Tholoth, M.; Li, Y.; Graham-Wooten, J.; Liang, Y.; Li, J.; Li, W.; Weiss, S.R.; Collman, R.G.; Bau, H.H. Single- and Two-Stage, Closed-Tube, Point-of-Care, Molecular Detection of SARS-CoV-2. Anal. Chem. 2021, 93, 13063–13071.
- Joung, J.; Ladha, A.; Saito, M.; Segel, M.; Bruneau, R.; Huang, M.W.; Kim, N.-G.; Yu, X.; Li, J.; Walker, B.D.; et al. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv 2020.
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874.
- Lucira Health Inc. Meet Lucira Check It. n.d.-b. Available online: https://www.lucirahealth.com/ (accessed on 28 October 2022).
- U.S. Food & Drug Administration. Authorization Letter Lucira COVID-19 All-in-One Test Kit. n.d.-h. Available online: https://www.fda.gov/media/143810/download (accessed on 28 October 2022).
- U.S. Food & Drug Administration. Authorization Letter Lucira Check-It COVID-19 Test Kit. n.d.-g. Available online: https://www.fda.gov/media/147492/download (accessed on 28 October 2022).
- Detect Inc. The Future of Testing Is in the Home. n.d.-b. Available online: https://detect.com/our-test (accessed on 28 October 2022).
- U.S. Food & Drug Administration, 2021c. Authorization Letter Detect COVID-19 Test. Available online: https://www.fda.gov/media/153663/download (accessed on 28 October 2022).
- Ali, Z.; Aman, R.; Mahas, A.; Rao, G.S.; Tehseen, M.; Marsic, T.; Salunke, R.; Subudhi, A.K.; Hala, S.M.; Hamdan, S.M.; et al. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020, 288, 198129.
- Yu, L.; Wu, S.; Hao, X.; Dong, X.; Mao, L.; Pelechano, V.; Chen, W.-H.; Yin, X. Rapid Detection of COVID-19 Coronavirus Using a Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP) Diagnostic Platform. Clin. Chem. 2020, 66, 975–977.
- U.S. Food & Drug Administration, 2021c. Authorization Letter Metrix COVID-19 Test. Available online: https://www.fda.gov/media/162401/download (accessed on 28 October 2022).
- U.S. Food & Drug Administration, 2021c. Authorization Letter DxLab COVID-19 Test. Available online: https://www.fda.gov/media/158977/download (accessed on 28 October 2022).
More
